Nov 2 2009
A new nuclear magnetic resonance (NMR) methodology being developed by Carnegie Mellon University's Roberto Gil has enabled a group of chemists to determine the correct chemical structure of a natural compound known as a withanolide, which has been shown to slow the growth of breast cancer cells. Gil's method can be used to determine the 3D chemical structure of small organic molecules, including natural products that exhibit medicinal properties.
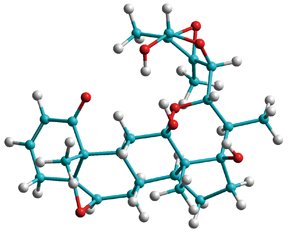
Chemists typically employ NMR spectroscopy to acquire information about the structure of chemical and biochemical compounds. However, the correct structural configuration of some compounds, like small organic molecules, is very difficult — and sometimes impossible — to reveal using conventional NMR techniques. Gil, associate research professor of chemistry and director of the Department of Chemistry's NMR Facility, has developed an NMR methodology to measure a relativity new NMR parameter known as Residual Dipolar Couplings (RDCs) in small molecules. Using RDCs, he was able to unambiguously determine the structure of the small molecule jaborosalactol 24, a naturally occurring steroid found in the Jaborosa parviflora plant. Researchers had unsuccessfully tried to elucidate the molecule's structure using conventional NMR techniques, but now, with the structure in hand, they are investigating the small molecule's function, and have found that it may have potential as an anti-cancer agent.
"It was impossible to figure out the compound's structure without RDCs," Gil said. "Conventional NMR techniques yielded three possible structural configurations for the compound. But RDCs revealed only one structure."
RDCs are readouts of the interaction between atomic nuclei induced to behave as tiny magnets in an NMR experiment. RDCs provide structural information about the molecule as well as the global orientation of all of the atoms in the molecule, even if the atoms are far away from each other. Traditional NMR methods cannot do this, according to Gil.
Over the past decade, RDCs have become an invaluable tool for determining the structure of large biological molecules, such as proteins and DNA, but the technique wasn't applicable to small molecules until recently. Small molecules need to be dissolved in an organic solvent before being analyzed in a typical NMR experiment. But to get a read out on RDCs, scientists first need to partially align the molecules that are tumbling around in the solution, which proved difficult. To overcome this problem, scientists — including Gil — began developing polymer gels that act like sponges that absorb the solution of small molecules. Gil, together with his Carnegie Mellon collaborators Nicolay Tsarevsky and Krzysztof Matyjaszewski, developed a gel called poly-methyl methacrylate (PMMA) that can be compressed or stretched, which causes the small molecules within the gel to partially align. This makes it possible to detect RDCs.
Although there are similar gels available, PMMA has a few advantages. During NMR experiments, the other polymer gels give off a background signal that is difficult to remove. With PMMA, the background signal from the polymer can be easily removed. Additionally, PMMA is easy to prepare and affordable.
"We are working to make this methodology easy and available to all chemists," said Gil, who is one of only a handful of scientists around the world currently using RDCs to determine the structure of small molecules. "Our technique helped a group of natural product chemists to unambiguously determine the correct configuration of a natural compound that has since shown anti-proliferative properties in a breast cancer cell line. Our technique can help anyone that has small molecules with difficult configurations."
In the current research, published in a recent issue of Angewandte Chemie, Gil worked with natural product chemists Manuela Garcia and Viviana Nicotra at the Universidad Nacional de Córdoba in Argentina; Armando Navarro-Vázquez from the Universidade de Santiago de Compostela in Spain, who developed the software to analyze RDCs; and Silvina Pagola from the College of William and Mary, who used powder x-ray diffraction to independently confirm the structure of jaborosalactol 24. This marks the first time that powder x-ray diffraction analysis has been used to determine the configuration of a complex natural product.