Nov 10 2009
A recently devised method of imaging the chemical communication and warfare between microorganisms could lead to new antibiotics, antifungal, antiviral and anti-cancer drugs, said a Texas AgriLife Research scientist.
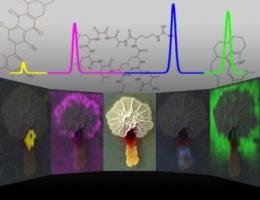 Bacteria produce many small molecules as both communication and warfare with other bacteria, but until new imaging techniques were recently developed, scientists only saw a small fraction of them, according to Dr. Paul Straight, Texas AgriLife Research scientist. In the center of this graphic is a photograph of two colonies of bacteria. On either side are images in false-color made with a mass spectrometer showing some of the molecules that are produced. At the top of the graphic are representations of the molecules. (Graphic by Jeramie Watrous, University of California, San Diego)
Bacteria produce many small molecules as both communication and warfare with other bacteria, but until new imaging techniques were recently developed, scientists only saw a small fraction of them, according to Dr. Paul Straight, Texas AgriLife Research scientist. In the center of this graphic is a photograph of two colonies of bacteria. On either side are images in false-color made with a mass spectrometer showing some of the molecules that are produced. At the top of the graphic are representations of the molecules. (Graphic by Jeramie Watrous, University of California, San Diego)
"Translating metabolic exchange with imaging mass spectrometry," was published Nov. 8 in Nature Chemical Biology, a prominent scientific journal. The article describes a technique developed by a collaborative team that includes Dr. Paul Straight, AgriLife Research scientist in the department of biochemistry and biophysics at Texas A&M University in College Station, Dr. Pieter Dorrestein, Yu-Liang Yang and Yuquan Xu, all at the University of California, San Diego.
"Microorganisms encode in their genomes the capacity to produce many small molecules that are potential new antibiotics," Straight said. "Because we do not understand the circumstances under which those molecules are produced in the environment, we see only a small fraction of them in the laboratory."
An example is the antibiotic erythromycin, which is often prescribed for people who are allergic to penicillin, Straight said.
"We know that Saccharopolyspora erythraea, the bacteria from which erythromycin is derived, encodes the capacity to produce numerous other small molecules that might be potentially valuable drugs," he said. "Conventional microbial culture and drug discovery techniques uncovered erythromycin. Other potentially useful metabolites may require some unconventional methods for identification."
Historically, medicinal drugs have been discovered serendipitously or by finding the active ingredient in homeopathic remedies, Straight said. For example, the use of blue mold for treating wounds was a folk remedy dates back to the Middle Ages. But scientists didn't isolate and purify the active ingredient, penicillin, until the early 20th century, which marks the beginning of the era of ‘natural product’ medicines originating from microorganisms.
Modern methods of drug discovery rely on screening technologies, knowledge of how infection is controlled and why diseases originate at the molecular level. Some new drugs can be designed accordingly from the ground up, often at significant cost, but serendipitous discovery of what nature has to offer is still a valid approach, he said.
Microorganisms, such as the bacteria that produces erythromycin, have been communicating and battling with each other for millennia using similar small molecules.
"What we learn about how microbes interact and exchange chemicals, and how the presence of one signaling molecule or antibiotic changes the output of potential antibiotics from a neighboring microbe, will guide us to new strategies for boosting the number of potential therapeutic drugs from any given bacteria," Straight said.
The National Institutes of Health recognizes the need to boost development of new drug compounds, he said.
"Globally, there is a shortage of new antibiotics that are being discovered by pharmaceutical companies in the traditional way and an ever-increasing number of multiple drug-resistant pathogens and newly emerging pathogens," Straight said.
The method of Straight, Dorrestein and colleagues employed an instrument called a "matrix-assisted laser desorption/ionization mass spectrometer." The device ionizes part of the sample with a laser beam while a crystalline matrix prevents the bio-molecules from being destroyed.
The plate upon which the bio sample sits is moved during the scan, from which hundreds to thousands of spectra are collected. The data is then processed as a grid and rendered as false-color by computer, then overlaid on a visual image of the sample.
Straight, Dorrestein and colleagues used two common bacteria that are cultured in the laboratory for their tests, Bacillus subtilis and Steptomyces coelicolor, both commonly found in soils. The bacteria were cultured together and their complex chemical interaction recorded using the mass spectrometer.
In competition for resources, the bacteria produced small molecules that alter antibiotic production from patterns present when cultured separately, Straight said. For example, they found that production of an antibiotic that targets Gram-positive organisms (Streptococcus and Staphylococcus are examples of Gram-positive organsims) was inhibited in one bacteria by the other.
The data reveal the chemical complexity of interspecies encounters. Using genetic sequencing, the researchers found that bacteria may dedicate up to 20 percent of their DNA to the bio-synthesis of small molecules in their communications and chemical battles with other microorganisms.