A team of researchers from the Yale University and Oak Ridge National Laboratory (ORNL) has developed nanopores with a radio-frequency electric field that can trap segments of biomolecules such as DNA.
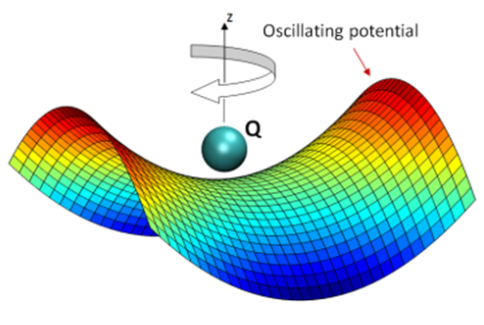 This representation of the Paul trap function illustrates how the oscillating electric field traps a particle.
This representation of the Paul trap function illustrates how the oscillating electric field traps a particle.
This technique for high-speed genomic sequencing device shows promise in bringing down the cost of human genome sequencing. This work is part of an initiative by the National Institutes of Health’s National Human Genome Research Institute to promote research on reducing the cost of human genome sequencing.
The research team has reported its hypothesis, computation and experimental results in a paper titled ‘Tunable Aqueous Virtual Micropore,’ published in the journal, Small. The team demonstrated that a charged nano or micro particle like a DNA segment can be trapped in an aqueous virtual pore. The water enabled a stable environment to maintain the integrity of DNA, while the virtual walls let the DNA to traverse the nanopore without interplaying with physical walls.
The research team was able to manipulate the stability and size of a virtual nanopore by applying external electric fields. This was not possible with a physical nanopore. The team formed the aqueous nanopore embedded in water on the basis of a linear Paul trap that detains particles in the presence of an oscillating electric field. It also experimentally demonstrated the trapping functionality of the aqueous nanopore by proving water’s capability in stabilizing trapping mechanisms.
Project Director Predrag Krstic informed that since a single DNA polymer is passed through a synthetic nanopore, the team reads electric signals that detect DNA bases by physically detecting single molecules. If the low-cost technique becomes achievable, then genomic sequencing can be utilized in daily clinical treatments, Krstic concluded.
Source: http://www.ornl.gov