Jul 19 2017
It is now possible to detect traces of biomolecules such as DNA with the help of a new “dynamic” technique based on the observation of association and dissociation events of gold nanoparticles.
The presence of the desired DNA sequence will result in reversibly binding together two nanoparticles. A report published in the journal Angewandte Chemie explains that this method is capable of differentiating true signals from noise and also detecting deviations of independent bases.
 Copyright Wiley-VCH
Copyright Wiley-VCH
The detection and quantification of biomolecules at very small concentrations is increasingly vital for applications such as monitoring cancer treatment, highly sensitive tests for biological weapons, early and precise diagnosis and forensic investigations.
The polymerase chain reaction (PCR) is the current method of choice and is based on the enzymatic replication of DNA. The false positives that could result from the tiniest amounts of impurity are considered to be a disadvantage of this method.
A new method for detecting extremely small amounts of DNA, without false positive results, signal amplification, or replication has presently been developed by Scientists working with Jwa-Min Nam at Seoul National University (South Korea). This method is based on the detection of independent binding events.
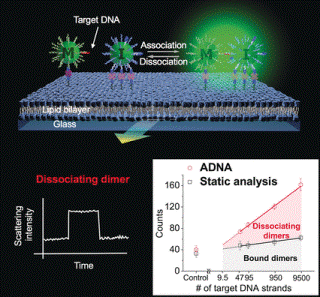
The binding partners constantly separate and then bind again resulting in the number of detectable results being multiplied and unspecific signals minimized. This associating and dissociating nanodimer analysis (ADNA) is based on the measurement of the scattering of light by gold nanoparticles with the help of dark-field microscopy.
Two types of gold nanoparticles and the sample are placed onto a glass slide coated in a lipid double layer. One type of gold nanoparticle contains binding sites on the surface that attach to the lipid layer. The other type of nanoparticle binds to the lipid layer, continuing to be mobile.
Both nanoparticles comprise of short single-stranded DNA segments that are corresponding to two varied sequences in the target DNA in order to bind it. When a mobile nanoparticle comes very near to an immobilized one, the target DNA will be able to bind them into a dimer.
The binding of two nanoparticles leads to coupling of their vibrations (plasmons). This modifies the color and intensity of scattered light, which can be identified in real time. The key to the accurate differentiation between the presence and absence of the target DNA refers to the dynamic analysis of dimers that dissociated during observation. The kinetics of the dissociation is majorly different for DNA with a single altered base and DNA that is an ideal match.
It was possible to specifically detect and consistently quantify ultra-low concentrations of the target DNA even in the existence of other DNA, such as in a sample of human blood serum. The detection limit was about 46 DNA copies under the test conditions used.