A new laser-based nanoparticle tracking analysis system is now available which allows nanoscale particles such as viruses and virus aggregates to be directly and individually visualized in liquids in real-time, from which high-resolution particle size distribution profiles can be obtained.
The technique is fast, robust, accurate and low cost representing an attractive alternative or complement to existing methods of nanoparticle analysis such as Dynamic Light Scattering (DLS), Photon Correlation Spectroscopy (PCS) or Electron Microscopy (EM).
By simultaneously and directly measuring the diffusion coefficient of individual particles, the dedicated Nanoparticle Tracking Analysis (NTA) software suite uniquely allows the user to automatically count and size the viruses and aggregates in a sample. Results are displayed as graphs of size against the count of individual particles (or size vs. relative brightness). This particle-by-particle approach overcomes the limitations inherent to other particle analysis systems which only generate mean particle size distribution data.

Figure 1. Typical virus image produced by the NanoSight instrument.
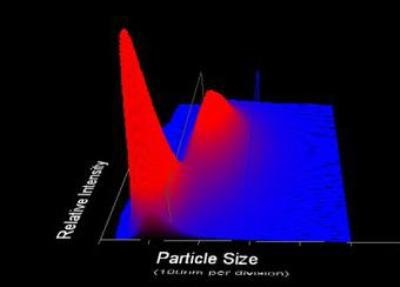
Figure 2. 3-Dimensional plot of particle size vs. relative particle intensity vs. particle count.
Vaccine Production/Development
Viral vaccine preparations must be proven (validated) to be both stable and to contain known proportions of active elements. NanoSight allows an immediate and direct estimation of product purity and concentration.
Similarly, the degree and rate of formation of aggregates in a virus preparation can be simply estimated using NanoSight allowing the manufacturer to develop improved product manufacturing processes and to optimize product shelf life.
As NanoSight allows all particles in the preparation to be visualized and sized, additional information about nanoparticle content is made available in a shorter time than would be available through conventional bio TCid50 or plaque assays.
For instance, the presence of larger particles (which NanoSight can both size and count) could represent either non-viral cell debris from the cell culture process or aggregates of virus particles containing many individual virions. In either case, such aggregates/contaminants represent a possible problem to the manufacturer, one which can be immediately identified with the NanoSight system.
Virus Clearance Studies
Virus clearance is assayed (validated) with high titre spikes of viruses (of various types depending on application). These are then used to challenge process steps which need to be qualified as a clearance step.
The data must be obtained with pure, non-aggregated virus spike material which must first be prepared at high titre and stored before use as the spike. NanoSight is ideally placed to establish the status of aggregation both before and after storage.
Virus Purification
The ability of NanoSight to rapidly establish the degree to which a virus preparation contains contaminants or aggregates and to be able to quantify the levels of such has proved invaluable to process developers interested in optimizing purification protocols for virus preparation.
For example, Figure 3 shows the difference between a partially purified virus preparation (white line) and the same sample having been successfully passed through an efficient purification protocol which effectively removed all contamination or aggregated material (red line). Note that the vertical axis represents particle concentration (virus particles/ml).
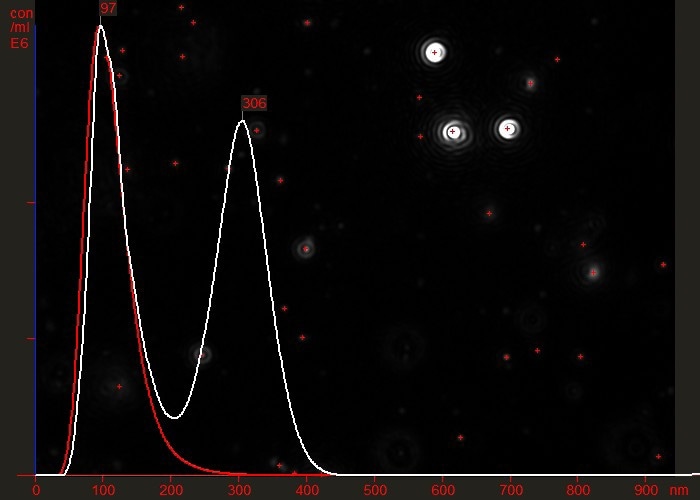
Figure 3. Overlaid particle size distribution plots of a virus preparation before (white) and after (red) a final purification step.
Bacteriophage-Based MRSA Protection – Phage Therapy
Virus particle detection and counting using NTA is providing essential information for researchers at the University of Strathclyde’s Institute of Pharmacy and Biomedical Sciences (IPBS). This team develop methods to employ naturally occurring bacteria to combat MRSA.
MRSA (or Methicillin Resistant Staphylococcus Aureus) is a variation of the bacterium Staphylococcus Aureus which has developed resistance to most antibiotics making it difficult to treat and potentially deadly. Whilst the “Superbug” can be killed with detergents, detergent dilution and application is often inconsistent and ineffective, making the bacteriophage route an attractive alternative.
It is in characterizing bacteriophage cultures that NanoSight is employed within a team lead by Dr Mike Mattey, Honorary Lecturer at IPBS. Prior to deploying bacteriophages as a dry-coat to protect high-risk bacterial invasion sites (sutures, instruments and wounds), the cultures need characterizations and their concentration needs assessing. NanoSight allows the team to view and size viral cultures rapidly in real-time and at low cost.
“The characterization of virus populations requires assessment of aggregation in the 20 nm – 1,000 nm range,” says Dr Mattey. “NanoSight provides fast and easy quantitative sample characterizations not possible with other methods and at a much lower cost. Additionally, NanoSight’s technology provides a reassuring view of the particle population that supports the counting results.”
Measurement Parameters
-
Sample pre-treatment is minimal requiring only dilution to 107 – 1010 per ml.
-
Accurate and reproducible analyses can be obtained from a 30 second analysis. Size distribution is a number distribution and hence sample concentration can be derived.
-
Real-time video clips allow time dependent phenomena such as aggregation or dissolution to be followed both qualitatively and quantitatively. Changes in both size and concentration can be calculated in such processes.
-
The minimum detectable size measurable depends on the particle type but most viruses and phage types can be seen, sized and counted. The absolute limit for the technique is 10 nm but is only possible for highly retractile materials.
-
The technique is absolute, requiring no calibration.
Virus Types
A wide range of virus types have been successfully analyzed to date and include:
- Adenovirus
- Cytomegalovirus (MCMV)
- Porcine papillomavirus
- Lambda phage
- Avian flu stimulant (TMV)
- Murine Leukaemia virus (MULV)
- Japanese Encephalitis virus (JEV)
- Cyanovirus
- Herpes Simplex
- Influenza
- Baculovirus
- M13
- M14

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information please visit Malvern Panalytical.