Nanoparticle Tracking Analysis (NTA) provides information on particle size distribution of samples in liquid suspension using the properties of Brownian motion and light scattering.
When particles in suspension are exposed to a laser beam traversing the sample chamber, they scatter the light, which makes them visible through a 20x magnification microscope. The camera mounted on this microscope captures the particles’ Brownian motion within its field of view (FOV) of roughly 100µm x 80µm x 10µm as a video file (Figure 1).
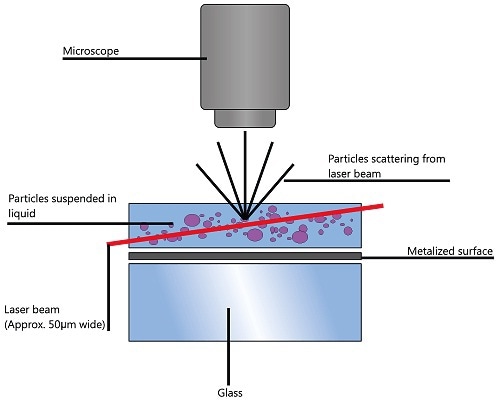
Figure 1. Schematic of the optical configuration used in NTA.
The center of the particles is identified by the proprietary NTA software to determine the average distance travelled by each particle in the x and y directions. From this value, the particle diffusion coefficient (Dt) can be calculated. When the solvent viscosity (η) and sample temperature (T) values are known, the sphere-equivalent hydrodynamic diameter (d) of the particles can be determined with the help of the Stokes-Einstein equation:
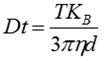
Where, KB = Boltzmann's constant.
NTA sizes each particle one at a time regardless of the other particles. Figure 2 illustrates the size distribution profile generated by NTA. Since the particles’ motion is tracked within a fixed FOV (roughly 100µm by 80µm) irradiated by a laser beam of roughly 10µm in depth, the sample’s scattering volume can be calculated. The particle concentration within this FOV is measured and extrapolated to a larger volume to estimate the concentration in particles per mL for an overall total or any given size class.
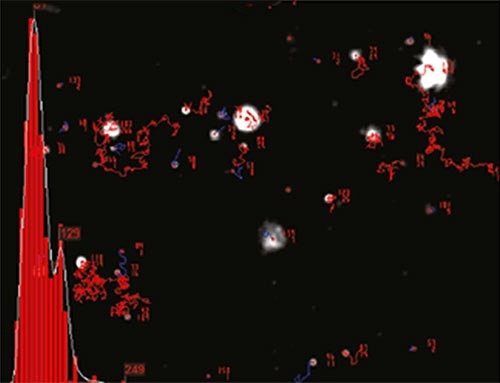
Figure 2. An example of the size distribution profile generated by NTA. The modal size for this sample is found to be approximately 70nm, with larger sized particles also present.
Advantages of NTA
NTA can visualize, size and enumerate nanoparticles in suspensions based on an individual basis. In addition, concentration and direct number frequency based particle size distribution profile can be determined using NTA. These capabilities of NTA make it suitable to perform in-depth analysis of virus preparations.
In the manufacture of vaccines, knowing the size of any phage or virus particle is often secondary to the virus particle concentration measurement and extent of aggregation. Hence, NTA is a suitable method for virus preparations, thanks to its ability to measure virus concentration by direct visualization regardless of virus infectivity. Likewise, NTA is capable of detecting, sizing, and measuring concentration of all except the smallest viruses, making it ideal over existing techniques used in the characterization of viruses and viral vaccines.
Virus Characterization, Sizing and Concentration Measurement
Manufacturers of viral vaccines must know the count of virus particles present in the preparation irrespective of virus infectivity and the degree of virus aggregation, which limits the shelf life of the final product. For this purpose, NTA has been evaluated. The results of an assessment of NTA involving the measurement of the count of both artificial and natural nanoparticles like adenoviruses concluded the supremacy of the NTA system over mathematical calculation and spectrophotometer techniques. However, the potential limitation of NTA was the concentration range over which it was applicable.
NTA was proven effective in a myriad of fields, including vaccine production, drug delivery, nanoparticle synthesis and aggregation. Another study involved the determination of total virus particles using NTA and compared the NTA results with the results of hemagglutination (HA) and end point dilution assay to determine total and infection virus particle count, respectively. The NTA system performed rapid virus concentration measurement in different samples and helped understanding the sample status pertaining to the degree of virus aggregation better.
In a comparison study of NTA, traditional plaque assay (PA) and quantitative polymerase chain reaction (qPCR) to detect three types of phages, NTA proved to be rapid and provided better performance in a comparatively clear medium over a maximum concentration range without any additional reagents, although is more capital expensive.
After optimizing a NTA-based method for phage, it can be reproduced in different labs, with an accuracy equivalent to PA carried out by different analysts but at a faster rate. This makes NTA suitable for future fundamental and applied research with bacteriophage. NTA also has the ability to visualize, size, and measure aggregates. In addition, NTA was also able to quantify fluorescently labeled virus particles, making it suitable to harvest materials, for which viruses must be differentiated from background host-cell debris and proteins.
In a study, NTA was used to corroborate the quantitation of intact oncolytic Vaccinia virus particles by means of viral quantitative capillary electrophoresis (Viral qCE). In another study, NTA was used to estimate the binding of DNA aptamers to vesicular stomatitis (VSV) and Vaccinia virus. The study results revealed the advantage of using NTA for all further advances of biomolecular binding analyses and aptamer assortment.
Vaccines and Virus-like Particles
Several research teams have reviewed the application of NTA in vaccine research recently. A research group conducted a study with an aim of developing large scale production protocols of virus-like particles (VLPs), which are potential candidates for new line of vaccine strategies. The researchers developed a simple, rapid and economical quantitation assay, which holds potential in the advancement and improvement of bioprocessing strategies for Gag-based VLPs.
Using NTA technology, another research group obtained virus particle measurements in liquids in real time and with no contamination or particle aggregation (clustering). The results allow the researchers to explore and manipulate the size, concentration, degree of aggregation of synthetic plant virus particles and tiny spherical animal and plant viruses. The rapid measurement capability of NTA was demonstrated in another study involving the characterization of biopharmaceuticals and procedures to determine size distributions.
Phage and Virus Templating
The results of a study involving the determination of phage concentration using NTA concluded the necessity to have a more realistic theoretical method to plaque formation to gain insights into the complex reactions between phage and its bacterium host in a spatially limited environment.
NTA enables simultaneous measurement of the average scattering intensity of a particle as well as its size based on its dynamic behavior. This capability allows discerning particles of the same size but with different refractive index by plotting these two values as a function of each other. As a result, virus particles of highly defined structure and homogenous size can be used as templates to manufacture highly monodisperse metallized particles using an electrodeless deposition metallization process.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information please visit Malvern Panalytical.