Sponsored by Nanosurf AGReviewed by Olivia FrostJan 4 2023
Over the last century, humans have experienced multiple severe viral outbreaks, including the Spanish flu and other influenza epidemics, HIV, SARS-CoV, MERS, Ebola, and the recent SARS-CoV2 pandemic. During this time, science has made significant progress in understanding the various facets of virus outbreaks, such as viral infectivity, pathogenicity, virulence, transmission, and the likelihood that these circumstances will result in an epidemic/pandemic.
Modern techniques have helped researchers to understand the virus life cycle better. Among these methods are viral replication and the virus-host interaction that includes cell culture, infectivity assays, polymerase chain reactions, fluorescence microscopy, electron microscopy, and immunoassays. Researchers now have improved knowledge of the structure and function of the viral particle, viral genome, RNA or DNA expression levels, and the virus-host cell interaction and immune response.
As a result of the progress and advances, newer vaccines and drugs have been developed to treat several different viruses. Atomic force microscopy (AFM) is one of the more recent techniques available for virus research. AFM is a cantilever-based technique that applies a sharp tip to examine surfaces at resolutions far below the optical diffraction limit. AFM also has applications beyond imaging, as it is a powerful tool for nano-mechanical probing and nanomanipulation.
Delivering a comprehensive toolbox of applications, AFM can address the structure, function, and host interaction of viruses. One of the main benefits of AFM, in contrast to electron microscopy, is that it can also function on samples immersed in liquid. This makes experiments on living cells or cell organelles at physiologically relevant conditions easy to conduct.
AFM imaging has been applied in the study of functional/infective virus particles, and to investigate the dimension and morphology and packaging of viral genomic material. Beyond imaging, AFM has found use in manipulating single viruses by force spectroscopy to examine the early events of virus-host interactions closely.
This article describes the process of high-resolution imaging and force spectroscopy on virus capsids attached to glass substrates and isolated cell nuclei.
Two cantilever-based approaches are presented for studying virus-host interactions at the single-cell level. In the first example, a hollow cantilever was used to deposit virus particles on a single cell to study the host cell response upon virus infection.
In the second example, single cells bound to a cantilever were infected with a virus. Over the course of several hours, the effect of an infection on the cell mass was carefully monitored.
High-Resolution Imaging and Force Spectroscopy of Herpes Simplex Virus 1 Capsids
AFM’s ability to observe samples in aqueous environments enables the observation of proteins, nucleic acids, and assemblies in a state as close to physiological as possible. Namely, ensuring samples such as Herpes simplex virus 1 (HSV-1) capsids remain fully hydrated enables observation of their unperturbed structure and study of their nanomechanics.
WaveMode imaging, a photothermally driven off-resonance operation mode, was employed to visualize HSV-1 capsids. Adsorption of the HSV-1 capsids to a modified glass surface led to capsids attaching insufficiently, as the surface area of the capsids interacting with the substrate is relatively small due to their shape. Consequently, imaging requires calm conditions to avoid displacing the capsids during the imaging process.
WaveMode, with its gentle imaging conditions, facilitates reproducible and fast visualization of HSV-1 capsids due to the minimal vertical and lateral forces acting on the capsids during imaging.
WaveMode imaging reveals the basic structural features of HSV-1 capsids, including their size or geometry.
Figure 1A displays AFM images of two HSV-1 capsids, which exhibit their overall icosahedral structure and the arrangement of capsomeres within the various facets of the icosahedral structure.
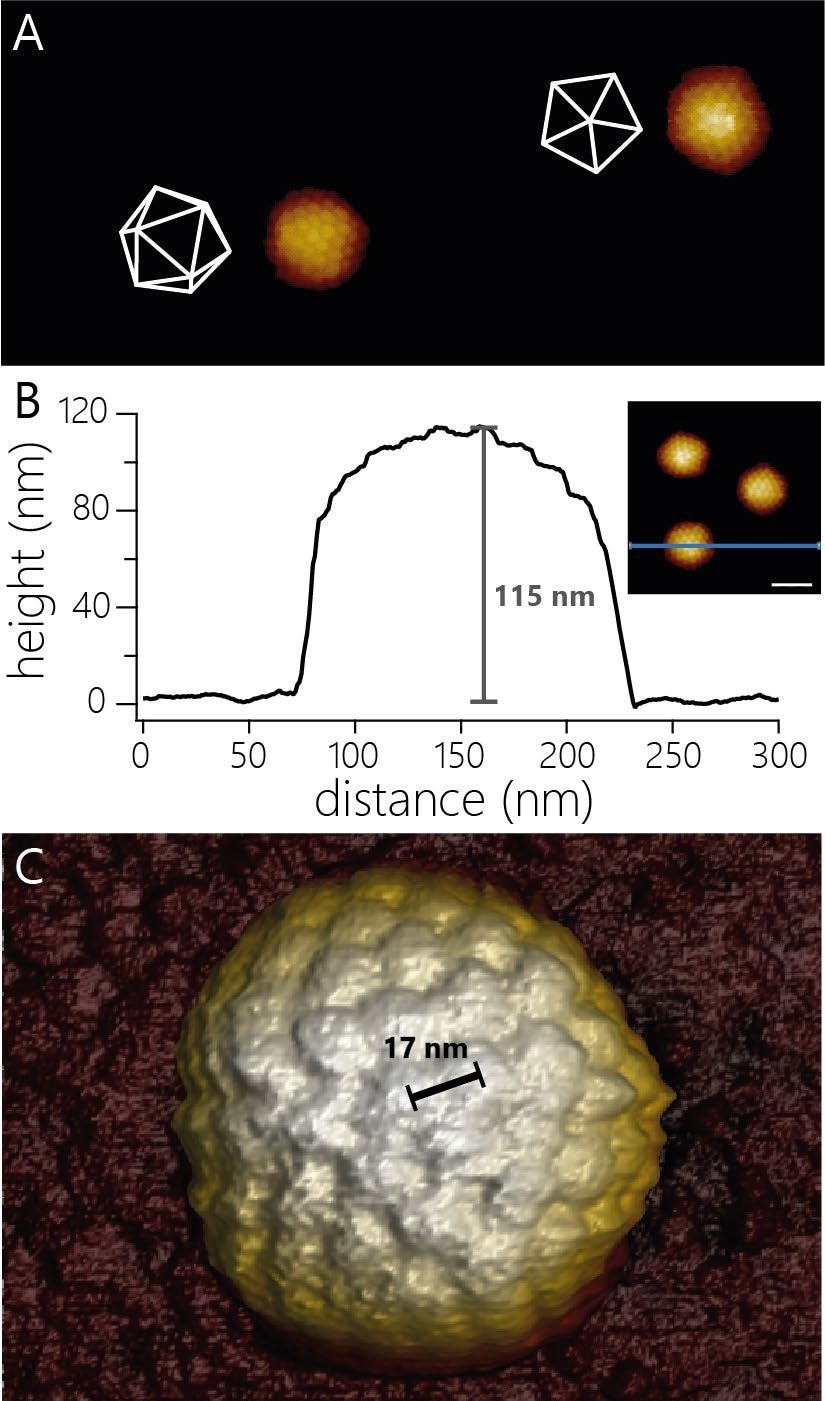
Figure 1. A) AFM topography of two virus capsids adsorbed to a modified glass surface. The capsids adsorbed in different orientations. The white wireframes indicate the surfaces of the capsids facing upwards. Image width 940 nm, color scale 55 nm. B) Cross-sectional profile of one of the three capsids shown in the inset (scale bar 100 nm). The capsid shows an apparent height of 115 nm. C) 3D render of AFM topography of a virus particle at higher resolution than in A). The individual capsomers can be observed and the center-to-center distance determined (17 nm). Image Credit: Nanosurf AG
The high resolution of the AFM also means that measuring the diameter of such virus particles is more precise.
For HSV-1, due to the icosahedral structure, the measured height is contingent on the orientation in which the particles adsorbed to the surface.
The examples displayed in Figure 1B show a height of 115 nm. At slightly higher resolution, it is possible to resolve more details of the capsid structure. For instance, the center-to-center spacing of capsomers can be determined (~17 nm; Figure 1C).
After virus capsids have been located using WaveMode imaging, AFM mode can be used to probe individual capsids to obtain the nanomechanical properties of these capsids. The nanomechanical properties of virus capsids are dependent on the structure of the protein-based capsid and on whether the capsids contain DNA or not.1
To measure the nanomechanical properties of capsids, the cantilever is guided toward the top of the capsid to apply an increasing force. Such experiments produce force-distance curves, as displayed in Figure 2A. The force curves can disclose a vast array of information beyond the nanomechanics of the probed capsid.
After contact with the capsid is made (Figure 2A, left dashed line) and upon application of force, the capsid's deformation occurs mainly elastic manner, as the force-distance curve shows a linear increase. The spring constant of the capsid can be seen in the slope of the force-distance curve (Figure 2A, orange line).
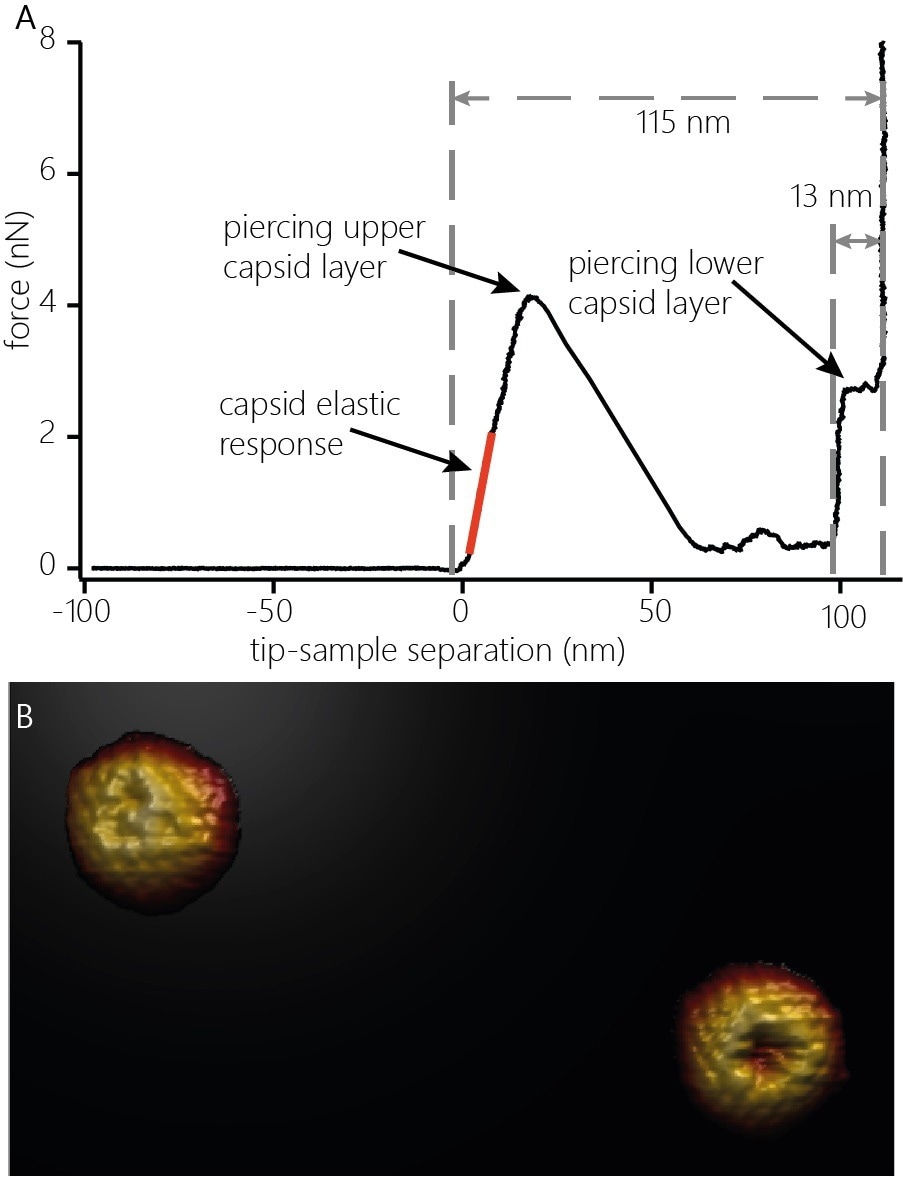
Figure 2. A) Indentation curve on an HSV-1 capsid. The curve shows the elastic response (stiffness, orange line) of the capsid upon application of force, the penetration of the upper protein layer and the penetration of the lower capsid layer. The distinct features of the force curve also allow estimating the height of the capsid (115 nm) and the thickness of the protein layer (13 nm). B) 3D render of two capsids after indentation. The capsids were indented with the cantilever until both capsid layers were penetrated and re-imaged. The defect caused by indentation as well as capsomers not directly affected by the indentation can be clearly resolved. Image width: 705 nm. Image Credit: Nanosurf AG
At some point, the capsid structure is compromised and fails due to the increased force applied to the capsid. The failure point is a sudden decrease of the force in the force-distance curve. The failure occurs locally rather than causing complete capsid destruction: Re-imaging the capsid after penetration shows a defect at the location where the cantilever came into contact with the capsid (Figure 2B).
The force-distance curve also displays information relative to the size of the capsid and the thickness of the capsid’s protein shell. After the first layer of proteins of the capsid has been pierced, the tip is further guided toward the surface and eventually comes into contact with the shell's inside at the bottom of the capsid (Figure 2A, middle dashed line). Further movement loads the lower protein layer of the capsid from the inside.
When the appropriate force is applied, the protein layer is pierced, and the tip pushes through and interacts with the underlying glass substrate (Figure 2A, right dashed line). The distance measured between the initial contact point with the capsid and the position at which the tip interacts with the glass surface shows that the capsid height is at the indentation point (~115 nm). The distance measured between contact with the lower protein layer and the interaction with the glass surface determines the overall thickness of the capsid shell (~13 nm)
Imaging of HSV-1 Capsids Bound to Isolated Cell Nuclei
In vivo, post-entry into the cell HSV-1 capsids attach to nuclear pore complexes (NPCs) on the outer surface of the cell nucleus to release their DNA into the cell nucleus. Evilevitch and Tsimtsirakis developed a reconstituted virus-nucleus system that facilitates the evaluation of the nanomechanical changes that occur when conducting DNA injection and reorganization of chromatin inside the nucleus.2
For the first time, WaveMode revealed high-resolution topographic information of HSV-1 capsids on intact nuclei and not just widely dispersed, flat nuclear membranes. The AFM images show individual HSV-1 capsids bound to NPCs and NPCs that do not contain capsids. (Figure 3A). With WaveMode imaging, capsids bound to the soft nuclei can be easily recognized by the individual capsomers that can be seen (Figure 3B).
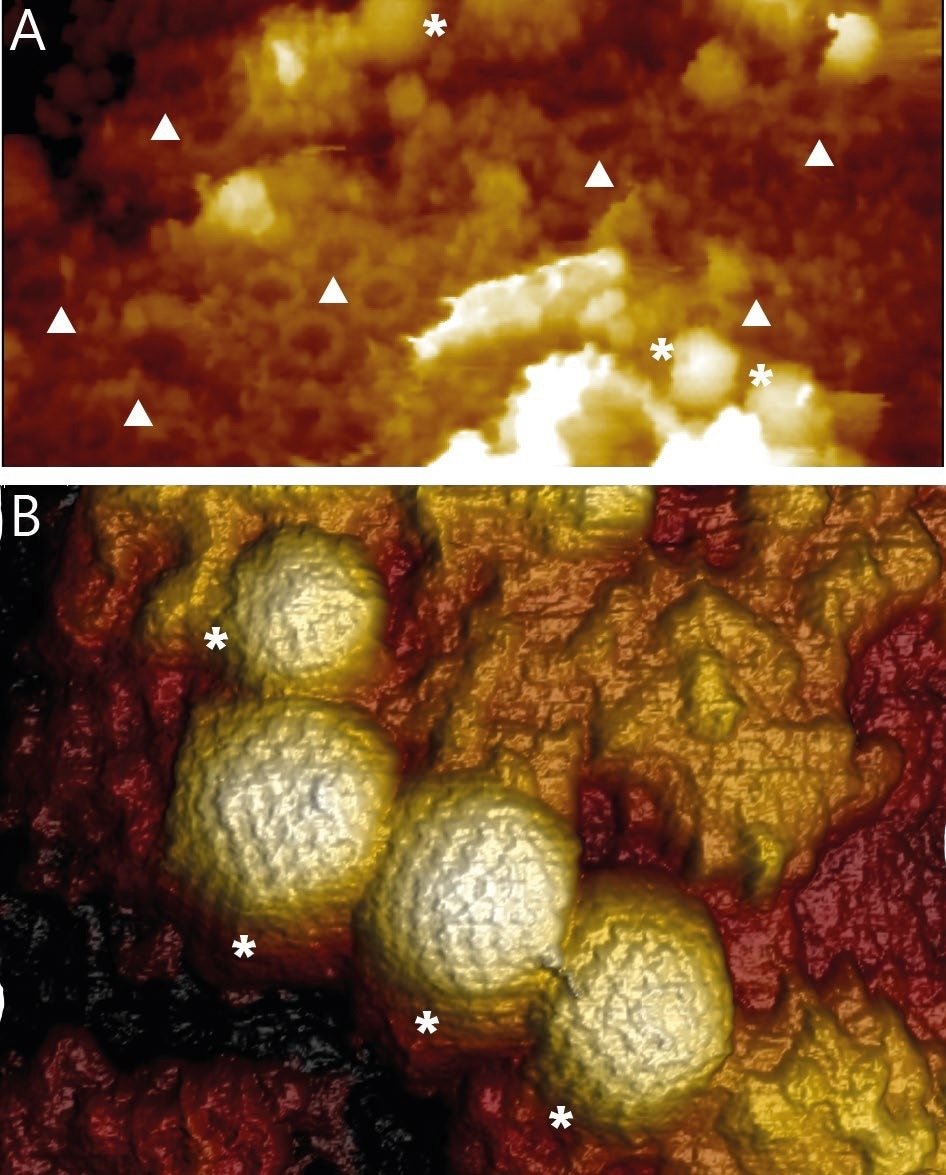
Figure 3. A) Overview image of the surface of an intact isolated and fixed nucleus. The image reveals NPCs (▲) and HSV-1 capsids (*) bound to NPCs. Image width 1650 nm. B) 3D rendered AFM topography of multiple HSV-1 capsids bound to an intact nucleus. Image width 590 nm. Image Credit: Nanosurf AG
The reconstituted virus-nucleus system enables imaging of the NPC-bound HSV-1 capsids and reveals the nanomechanical properties of nuclei under various conditions.
Figure 4A exhibits an indentation curve representing a nucleus incubated with DNA-containing HSV-1 capsids.
Repeated indentation of several capsids reveals Young’s modulus of 214 ± 10 kPa (average ± SE) for nuclei incubated with C-capsids (Figure 4B).
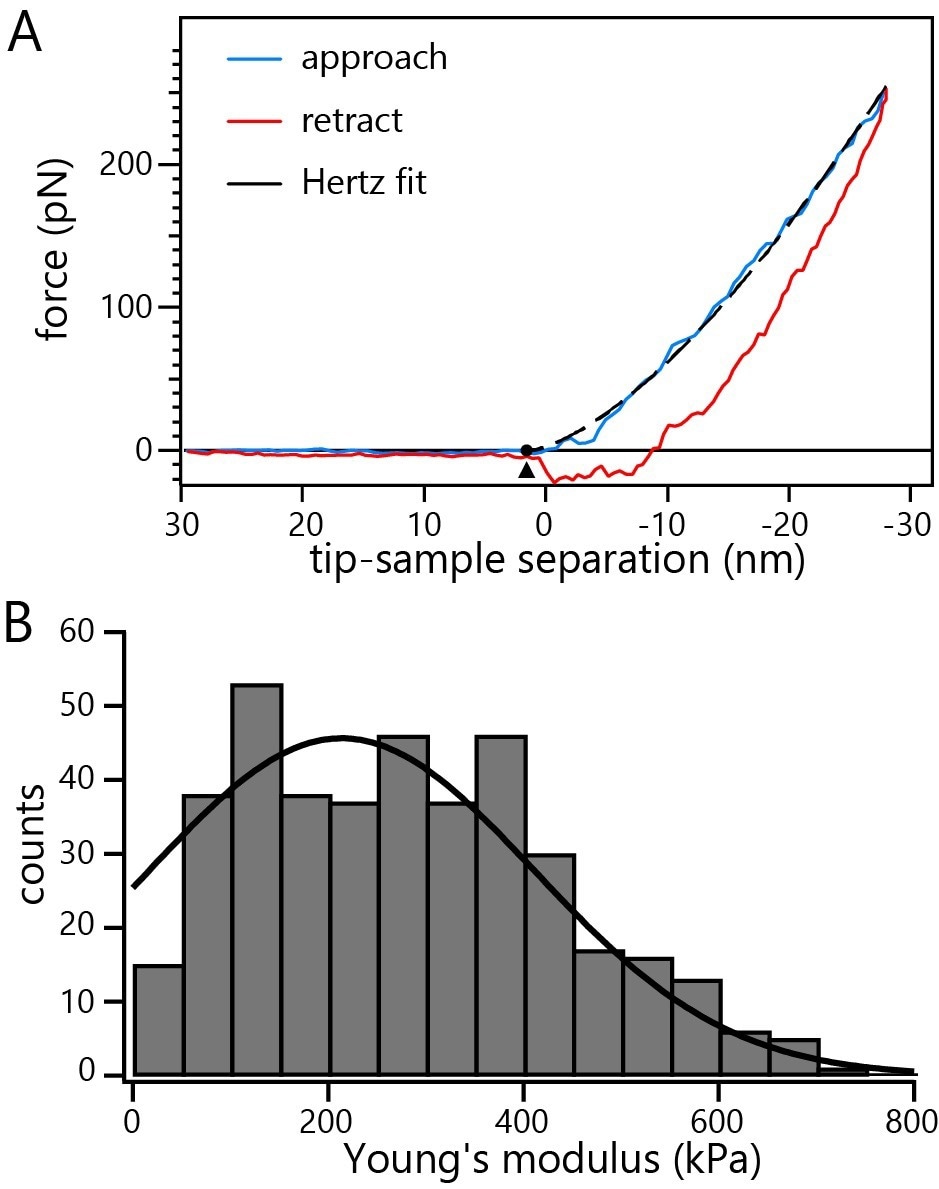
Figure 4. A) Representative indentation curve on a non-fixed nucleus incubated with C-capsids. The dashed line represents the fit of the Hertz model to the data. B) Histogram of Young’s moduli from repeated probing of multiple non-fixed intact nuclei incubated with C-capsids. Image Credit: Nanosurf AG
Cooperative Vaccinia Infection
AFM is not only applicable in imaging and spectroscopy applications but it can also be used as a manipulation tool. An example of this is the study evaluating the cooperativity of virus particles in cell infection to determine how the probability of virus infection is contingent on the number of virus particles attacking a cell.
To closely examine the cooperativity of virus infection, good, precise control is required on the timing of infection and the number of virions to which a cell is exposed.
Two innovative labs, the Laboratory of Biosensors and Bioelectronics and the Institute of Microbiology of ETH Zurich, used the FluidFM® to deliver a definitive number of virus particles onto single cells. FluidFM is a cantilever-based manipulation tool that can be introduced to AFM and utilizes hollow cantilevers to deposit or aspire material locally.3 A schematic of the FluidFM® technology is displayed in Figure 5.
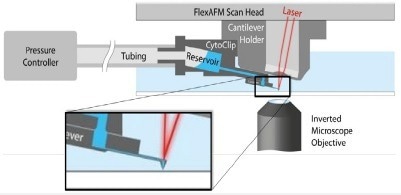
Figure 5. Schematic of AFM with FluidFM®. A hollow cantilever featuring an opening at its free end is filled with liquid from a reservoir. By applying a positive or negative pressure liquid can be secreted out of or aspired into the cantilever to locally manipulate a sample. The AFM handles accurate force control, and the complete system is integrated on an inverted, optical microscope to provide optical access to the sample. Image Credit: Nanosurf AG
In the study presented here, an AFM with FluidFM functionality was included with a fluorescence microscope to assess the single-cell response after exposure to different numbers of virus particles. The research groups at ETH developed a protocol to use FluidFM to deposit 1-12 mature vaccinia virions on single cells.4 The virus deposition process is outlined in Figure 6.
To ensure that the solution was not contaminated by the virus particles, which could cause uncontrolled infection, the FluidFM hollow cantilever was pushed gently onto the cell, creating a seal between the cantilever and the cell surface. Since the AFM delivers sub-nanonewton force control, the cells are not harmed during this process.
To follow the deposition of virions and the progress of the infection, the researchers utilized a recombinant vaccinia virus (VACV). This included the fluorescent protein mCherry into the virus core protein A5. This fusion protein facilitated the tracing of the assembled virion particles using fluorescence.
This enables counting the number of deposited particles and monitoring the subsequent assembly of new virions in the late infection phase. The virus also encoded for eGFP under the control of an early/late viral promoter. The eGFP was expressed after penetrating the host cells, creating a report of the various infection phases (Figure 7). The results of this study demonstrate the cooperativity of the virus infection.
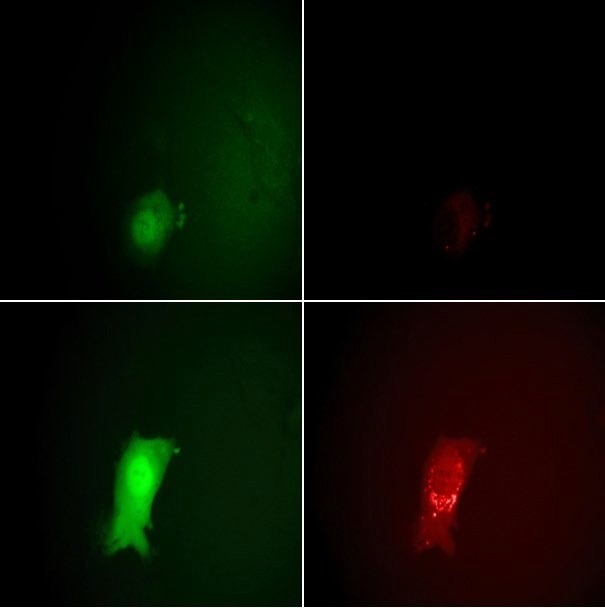
Figure 7. Monitoring the stages of the VACV lifecycle microscopically by eGFP and mCherry fluorescence signals. Top row: 7 hours post-infection, bottom row: 11.5 hours. The intensity of the EGFP signals on the left clearly show the entering of the early and late viral gene expression phases. The strong increase in the mCherry intensity at the right between 7 and 11.5 hours indicates the assembly of new virions. Image Credit: Nanosurf AG
A cell only had about a 10% chance of infection when targeted by a single virus (n=73). This probability increased to approximately 35% when two particles (n=23) were introduced and to 90% when attacked by four virus particles (n=20). The potential reason for this cooperativity is that the host cells' antiviral mechanism is compromised when multiple viruses arrive simultaneously.
Effect of Virus Infection on Cell Mass
In 2017, researchers at the Biophysics lab of ETH Zurich presented an inertial pico-balance to non-invasively measure the mass of adherent cells at high mass and time resolution.5
The PicoBalance possesses picogram mass sensitivity and millisecond time resolution. The mass derives from the change in resonance frequency of a cantilever, which is oscillated with subnanometer amplitudes via photothermal excitation.
There is a decrease in the cantilever’s resonance frequency when the cell is attached, and any further changes in the mass of a cell alter the resonance frequency of the cantilever accordingly. A schematic of the principles of the pico-balance is shown to monitor the cell mass for several days. The system is fitted with a special sample holder controlling humidity, temperature, and CO2, which acts as a miniature cell culture incubator. A schematic of this setup is displayed in Figure 9.

Figure 8. Schematic of the PicoBalance setup. The cantilever is oscillated at about 0.5 nm amplitude using an intensity-modulated laser at the base of the cantilever (dark red). Another laser focused on the free end of the cantilever (light red) is used to monitor the cantilever's resonance frequency. The mass of the cell attached to the cantilever and its change can be derived from the resonance frequency and its change. Image Credit: Nanosurf AG
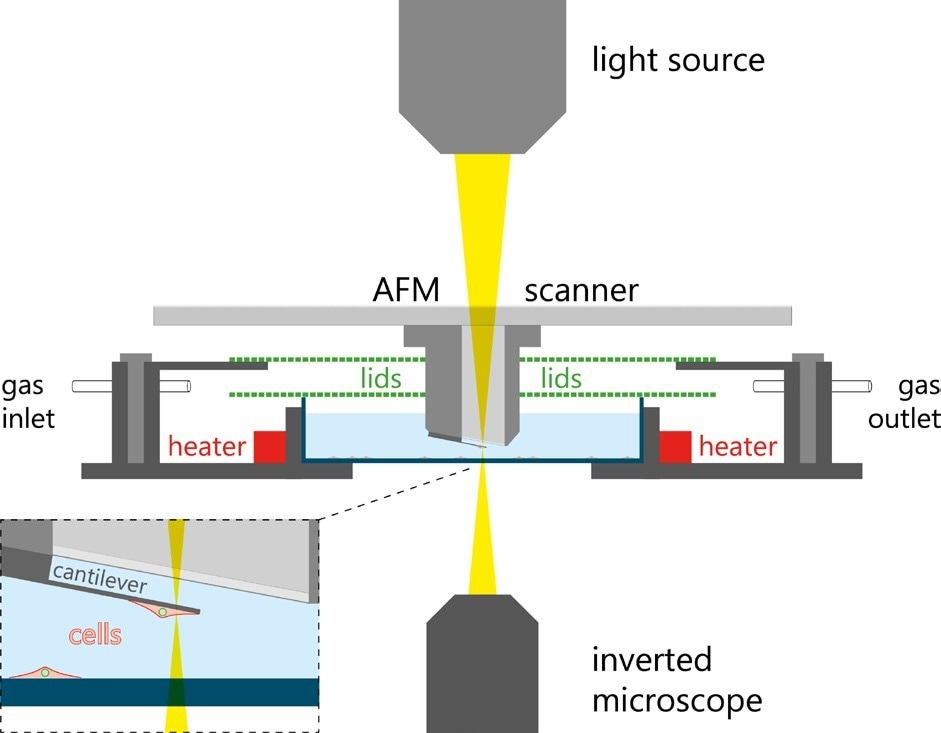
Figure 9. Schematic of the PicoBlance setup. Image Credit: Nanosurf AG
The investigators selected HeLa cells as a model system and introduced the cells to collagen-I-coated cantilevers. To assess the effect of a virus infection on the cell mass, contact between a single cantilever-bound HeLa cell was made with a vaccinia virus (VACV)-infected BSC 40 cells seeded on the bottom of the incubator cell. The cantilever-bound cell could then be infected selectively.
A recombinant vaccinia virus containing the eGFP-tagged core protein A4 was used to infect the cells to follow new virus particle production by fluorescence microscopy. Over time, the fluorescence intensity of the infected cells increased, suggesting that new virus particles were forming in the cells.
Both control and infected HeLa cells demonstrated that the mass of the attached cells fluctuated periodically by just a few percent every few seconds. However, the mass of the infected cells was constant over prolonged periods and did not enter mitosis. Control cells that did not come into contact with infected cells increased mass gradually and divided on the cantilever (Figure 10).
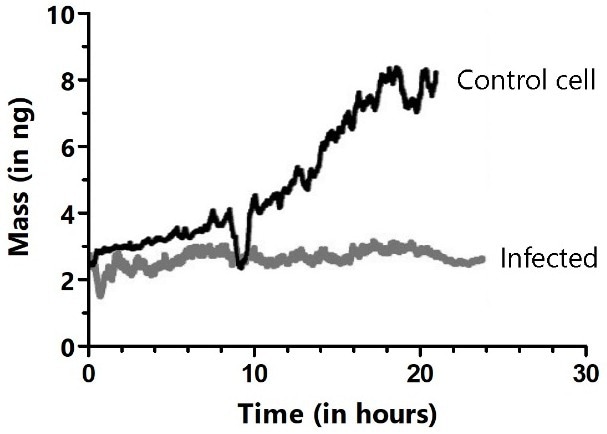
Figure 10. Long-term cultivation and mass monitoring of control (black trace) and virus-infected HeLa cells (gray trace). Cells were infected by bringing the cell attached to the cantilever in contact with an infected cell in the Petri dish. The dips in the black trace show where mitosis occurred, during which cells rounded up and adhered only weakly to the cantilever. Image Credit: Nanosurf AG
Conclusion
AFM is a powerful technique to analyze structure and nanomechanics and manipulate biological samples under physiological conditions. It can be combined with advanced optical techniques to allow correlation of optical characterization of the sample cells in parallel to the AFM experiments.
In this article, two applications were discussed regarding the study of the structure and nanomechanics of virus capsids that can be extended further towards analyzing the functional and mechanistic aspects of viruses and the virus-host interaction. Furthermore, two studies of host-virus interactions were evaluated at the single-cell level that surpass what AFMs are usually used for.
In the first instance, AFM was applied as a manipulation tool utilizing FluidFM technology. In the second case, the mass development of virus-infected cells was evaluated post-infection. Previous virus research has led to the development of new treatments and vaccines. New and emerging viruses such as the recent SARS-CoV2, which caused the COVID-19 pandemic, stress the importance of ensuring virus research improves to promote a faster response to outbreaks in the future.
References and Further Reading
- 1. Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure. N Martín-González, M. Hernando-Pérez, G.N. Condezo, M. Pérez-Illana, A. Šiber, D. Reguera, P. Ostapchuk, P.Hearing, C. San Martín, and P.J. de Pablo. Nucleic Acids Research 2019, 47(17), 9231–9242
- Reconstituted virus–nucleus system reveals mechanics of herpesvirus genome uncoating. A. Evilevitch & E. Tsimtsirakis. QRB Discovery 2022, 3, E2.
- Force-controlled manipulation of single cells: from AFM to FluidFM. O. Guillaume-Gentil, E. Potthoff, D.Ossola, C.M. Franz, T.Zambelli, & J.A. Vorholt. Trends in Biotech. 2014. 32(7), 381-388.
- Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. P. Stiefel, F.I. Schmidt, P.Dörig, P.Behr, T. Zambelli, J.A. Vorholt, & J. Mercer. Nano Letters 2012, 12(8), 4219-4227.
- Inertial picobalance reveals fast mass fluctuations in mammalian cells. D. Martínez-Martín, G. Fläschner, B. Gaub, S. Martin, R. Newton, C. Beerli, J. Mercer, C. Gerber & D.J. Müller. Nature. 2017. 550, 500– 505

This information has been sourced, reviewed and adapted from materials provided by Nanosurf AG.
For more information on this source, please visit Nanosurf AG.