Jan 29 2009
Much debate has surrounded the mostly unknown function of non-coding RNAs, which have recently been shown to constitute a surprisingly large proportion of gene transcripts, rather than encoding proteins. Now, as reported in Nature*, a research team led by Kunihiro Ohta of the RIKEN Advanced Science Institute, Wako, has discovered a new role for such non-coding RNAs in gene expression in the fission yeast, Schizosaccharomyces pombe.
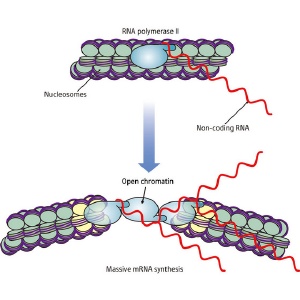 Conversion of chromatin from a closed to an open configuration.
Conversion of chromatin from a closed to an open configuration.
Most DNA is wrapped up in chromatin of the cell’s nucleus, which in turn is packaged in chromosomes. Chromatin can be in either a ‘closed’ or an ‘open’ configuration, affecting the accessibility of the DNA to proteins called transcription factors that regulate gene expression.
Ohta and colleagues showed that non-coding RNAs are involved in the step-wise conversion of chromatin from the closed to the open configuration during the induction of one particular gene in fission yeast. This supports some previous suggestions that non-coding RNAs could play an important role in regulating gene expression.
The gene studied by Ohta and colleagues is induced when yeast cells are starved of glucose. The most common transcript encodes the protein product of the gene. However, they also showed that three rarer, longer RNA transcripts are also produced, and that their expression is initiated far upstream of the main coding sequence of the gene.
As these longer RNA transcripts do not appear to have any special protein coding function, and the researchers set out to explore whether they have important functions during expression of the gene. They showed that the chromatin at the gene is progressively converted to an open configuration, as the non-coding RNAs are transcribed by RNA polymerase II.
Using various mutant yeast strains, the researchers then showed that transcription factors known either to induce or repress expression of the gene also regulate chromatin configuration in collaboration with the RNA polymerase.
Based on their findings, Ohta and colleagues propose a model for the regulation of the chromatin structure at the yeast gene.
“Transcription-mediated regulation of gene expression could also exist in humans,” say the researchers. Whether or not this turns out to be the case, the present study “demonstrates a new mechanistic role for non-coding RNAs in gene activation.”
The hunt is now on to discover the exact mechanism by which the non-coding RNAs affect the configuration of chromatin.
- Hirota, K., Miyoshi, T., Kugou, K., Hoffman, C. S., Shibata, T. & Ohta, K. Stepwise chromatin remodelling by a cascade of transcription initiation on non-coding RNAs. Nature 456, 130–134 (2008).