Dec 15 2008
Zackery Riley, BSc, MBA, is a senior research associate in the Methods Development group at the Allen Institute for Brain Science. Andreas Jeromin, PhD, is currently the manager of the Methods Development group at the Allen Institute for Brain Science. Their interest is focused on the development of novel and improved gene expression platforms and their integration with other high-throughput technologies.
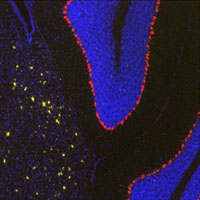 Shown is a multi-labeled Qdot image of Neuropeptide Y (NPY) and Calbindin 1 (Calb1) in a C57BL6/J mouse brain. Calb1 expression can be seen labeled in red (streptavidin-QD655) within the Purkinje layer of the cerebellum. NPY cortical expression is shown in Gold, (streptavidin-QD585).
Shown is a multi-labeled Qdot image of Neuropeptide Y (NPY) and Calbindin 1 (Calb1) in a C57BL6/J mouse brain. Calb1 expression can be seen labeled in red (streptavidin-QD655) within the Purkinje layer of the cerebellum. NPY cortical expression is shown in Gold, (streptavidin-QD585).
The Allen Brain Atlas, developed by the Allen Institute for Brain Science, is the first large-scale atlas of gene expression in the mouse brain, using chromogenic in situ hybridization (cISH) to detect the location of over 20,000 genes.
To overcome the limitations of a single label chromogenic cISH platform, Zack Riley, Emi Byrnes, Sheana Parry, Nick Dee and Andreas Jeromin developed a high-throughput multispectral quantum dot (Qdot) ISH platform (Fig 1). This new ISH tool provides a mechanism to systematically examine spatial gene expression patterns, aided by multispectral imaging, in the mouse and human brain.
The intrinsic photostability, brightness and tunability of Qdots is critical in providing superior signal-to-noise of the ISH signal and long-term stability, while minimizing photobleaching and allowing re-scanning of images. The photostability and tunability of the quantum dot nanocrystals is an ideal technology for examining localized and scattered gene expression in situ.
The team tells the NWN that they are currently extending the technology to incorporate additional labels for multiplexing to examine the spatial relationships between multiple genes in both two and three dimensions within a single tissue section. Combining Qdot technology and high-throughput automation will allow quick and efficient analyses of the unique characteristics of gene families and cell types, and will provide a molecular tool to understand their role in the normal and diseased brain.