Jul 27 2018
For the first time, researchers have been successful in measuring the attributive patterns of folds that give proteins their three-dimensional shape in water at the nanoscale. They achieved this by enhancing a technique they helped develop in the recent past.
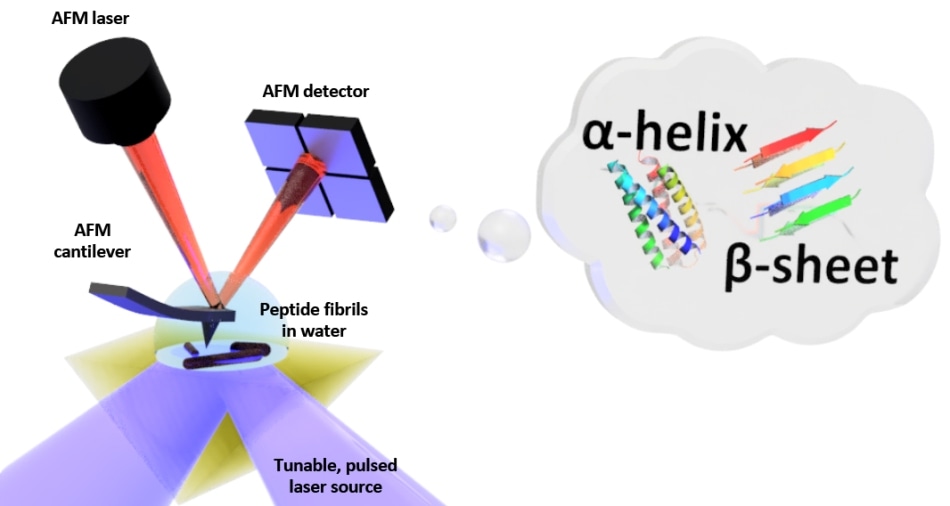 Schematic of the setup for photo-thermal induced resonance (PTIR), which includes an infrared laser source and atomic force microscope (AFM) cantilever with a sharp tip that touches the sample and vibrates in response to the sample’s light-induced expansion. PTIR can determine the folding pattern (called for example alpha-helix, beta-sheet) of peptides (amino acid chains) in water with nanometer-scale resolution. (Image credit: NIST)
Schematic of the setup for photo-thermal induced resonance (PTIR), which includes an infrared laser source and atomic force microscope (AFM) cantilever with a sharp tip that touches the sample and vibrates in response to the sample’s light-induced expansion. PTIR can determine the folding pattern (called for example alpha-helix, beta-sheet) of peptides (amino acid chains) in water with nanometer-scale resolution. (Image credit: NIST)
The technique developed by scientists from the National Institute of Standards and Technology (NIST) and their collaborators will help researchers understand the behavior of biomolecules in watery environments analogous to those in cells. These insights, on the other hand, could improve our knowledge of major diseases, such as Alzheimer’s, that are associated with “mistakes” in protein folding.
Life, as it is known, cannot survive if proteins do not fold into correct patterns resulting in sheets, helices, and other shapes that give proteins their three-dimensional structure. The precise shapes of proteins allow them to transfer oxygen, eliminate harmful bacteria, and carry out other important tasks in the body. Improperly folded proteins will not function and at times produce toxic fragments, such as those related to neurodegenerative disorders.
In order to gain insights into the finesse of folding, researchers must analyze in depth the arrangement of amino acid chains that are shorter and simpler compared to proteins—known as peptides—and how they fold, assemble, and rotate to form a range of shapes, or conformations. Biologists choose to investigate proteins and peptides immersed in water since that environment is in close approximation with the conditions inside living cells.
Methods like infrared spectroscopy developed earlier for determining the conformation of proteins lack the fine spatial resolution to analyze the tiny and diverse assemblies of misfolded and properly folded proteins. Moreover, these methods do not work well in an aqueous environment since infrared light is strongly absorbed by water, hindering the analysis. A pioneering method called photo-thermal induced resonance (PTIR) was also severely hampered by the presence of water. This method has recently allowed scientists to investigate peptide structure and conformation in air at nanoscale resolution.
At present, NIST scientists and their collaborators have shown that it is possible to adapt PTIR to acquire conformational structure at the nanoscale in water with the help of two chemically similar peptides called diphenylalanine and Boc-diphenylalanine. Diphenylalanine is related to beta-amyloid, a sticky, larger peptide associated with Alzheimer’s disease.
PTIR is a powerful technique that had already shown promise for the study of biological systems, but the possibility to use this with samples in a liquid environment will greatly improve its use in this area.
Georg Ramer of NIST and the University of Maryland in College Park
Ramer and NIST researcher Andrea Centrone, together with their collaborators at the University of Cambridge in England, reported their study in an article recently published online in ACS Nano.
PTIR ascertains the chemical composition of materials at nanoscale resolution by merging an atomic force microscope (AFM) with light from an infrared laser operating over a range of wavelengths. The attributive wavelengths of infrared light that are absorbed by the sample are similar to a molecular fingerprint, disclosing its chemical composition. The material heats up at every point on the sample at which infrared is absorbed, making it to quickly, but ever so slightly, expand. The sharp tip of the AFM that protrudes from a cantilever detects the expansion, which oscillates similar to a diving board every time the sample expands. The amount of light absorbed by the sample is directly proportional to its expansion as well as to the strength, or amplitude, of the oscillations.
Although PTIR is an efficient method, it is highly challenging to be used in a water environment. Infrared light is strongly absorbed by water, resulting in an absorption signal with the ability to interfere with efforts to determine the chemical structure of the sample. Furthermore, the drag force exerted by water is considerably stronger compared to that in air and it typically weakens the PTIR signal by strongly damping the oscillations of the cantilever of the AFM.
To restrict the absorption of infrared light by water, the researchers positioned a prism between the sample and the laser. The function of the prism is to restrict the infrared light to the surface of the sample, thereby reducing the amount of light that could leak out and interact with the water. In order to overcome the issue of damping, they used a laser with the ability to operate at frequencies of up to 2000 kHz. That allowed the scientists to match the frequency of the laser pulses to one of the higher frequencies at which the cantilever oscillates. Similar to pushing a child on a swing at just the correct interval, the frequency matching improved the amplitude of the oscillations of the cantilever, thereby partially offsetting the damping caused by water.
The researchers demonstrated the accuracy of their technique by comparing the PTIR measurements of diphenylalanine and other peptide samples in two environments: air and water. (Since the peptides folded the same way in both mediums, it was easier to carry out the comparison.) Astonishingly, the researchers accomplished similar spatial resolution and contrast in air and water, showing for the first time that it is possible to perform measurements in a water environment in an accurate manner, exhibiting the exact conformation of peptides with nanoscale resolution.
“This finding is important to biologists who want to understand protein structure and folding in environments as close as possible to those in cells,” stated Centrone.