Jan 14 2019
There are several occasions when human cells have to move. Formation of the body (embryonic development) is guided by mobile cells. Immune cells wander to capture undesirable intruders, and healing cells (fibroblasts) travel to repair wounds.
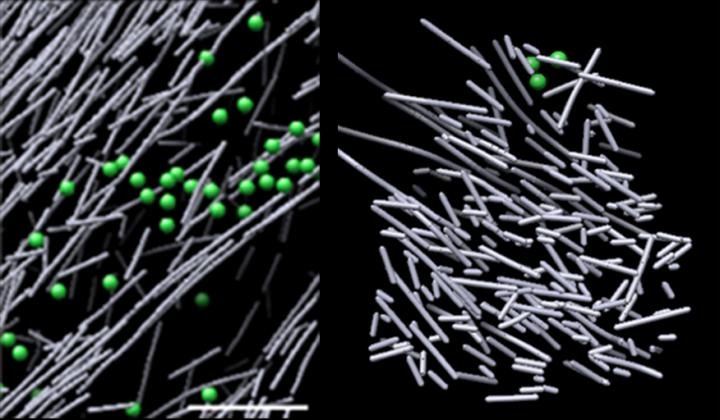 Cryo-electron microscope images of actin assembly in the cell in the absence of a molecular signal (left) and the haystack-like actin filament nanoscaffold that was induced in response to a molecular signal (Rac1) and promotes cell movement (right). The structure sprang into view in defined regions where Rac1 was activated, and quickly dissipated when Rac1 signaling stopped—in as little as two and a half minutes. (Image credit: Sanford Burnham Prebys Medical Discovery Institute)
Cryo-electron microscope images of actin assembly in the cell in the absence of a molecular signal (left) and the haystack-like actin filament nanoscaffold that was induced in response to a molecular signal (Rac1) and promotes cell movement (right). The structure sprang into view in defined regions where Rac1 was activated, and quickly dissipated when Rac1 signaling stopped—in as little as two and a half minutes. (Image credit: Sanford Burnham Prebys Medical Discovery Institute)
However, not all movement is advantageous. For instance, when cancer cells acquire the ability to move all around the body (metastasis), tumors become the most dangerous. Specific bacteria and viruses can exploit the motility machinery of the cells to attack human bodies. The key to learning how to stop or endorse the motility of the cells to improve human health is gaining insights into how cells move—and the rod-like actin filaments that drive the process.
Researchers from Sanford Burnham Prebys Medical Discovery Institute (SBP) and the University of North Carolina at Chapel Hill (UNC-Chapel Hill) have currently used one of the most powerful microscopes in the world to identify a dense, disorganized, and dynamic actin filament nanoscaffold—looking like a haystack—that is stimulated in response to a molecular signal. Scientists, for the first time, have directly visualized a structure at the molecular level, which is stimulated in response to a cellular signal—a key finding that widens the understanding of motility of human cells. The study has been reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Cyro-electron microscopy is revolutionizing our understanding of the inner workings of cells. This technology allowed us to collect robust, 3D images of regions of cells—similar to MRI, which creates detailed images of our body. We were able to visualize cells in their natural state, which revealed a never-before-seen actin nano-architecture within the cell.
Dorit Hanein, PhD, Professor, Bioinformatics and Structural Biology Program, SBP
Dorit Hanein is the senior author of the paper.
As part of the research, the researchers compared nanoscale images of mouse fibroblasts with time-stamped light images of fluorescent Rac1—a protein that regulates cell movement, response to force or strain (mechanosensing), and pathogen invasion—using SBP’s cryo-electron microscope (Titan Krios), artificial intelligence (AI), and custom-made computational and cell imaging techniques. This technically complex workflow—which bridged five orders of magnitude in scale (tens of microns to nanometers)—took years to grow to its present level of accuracy and robustness and was enabled through experimental and computational efforts of the structural biologist teams at SBP and the biosensors team at UNC-Chapel Hill.
The images showed a densely packed, scaffold-like, disorganized structure consisting of short actin rods. These structures appeared in definite areas where Rac1 was triggered and rapidly dissipated when Rac1 signaling stopped—in just about two and a half minutes. This dynamic scaffold had a stark difference with various other actin assemblies in regions of low Rac1 activation—some made up of long, aligned rods of actin, and others made up of short actin rods branching from the sides of longer actin filaments. The volume covering the actin scaffold completely lacked common cellular structures, such as microtubules, ribosomes, vesicles and so on, probably because of the structure’s intense density.
We were surprised that experiment after experiment revealed these unique hotspots of unaligned, densely packed actin rods in regions that correlated with Rac1 activation. We believe this disorder is actually the scaffold’s strength—it grants the flexibility and versatility to build larger, complex actin filament architectures in response to additional local spatial cues.
Niels Volkmann, PhD, Professor, Bioinformatics and Structural Biology Program, SBP
Niels Volkmann is the co-corresponding author of the paper and led the computational part of the study.
Further, the researchers desire to extend the protocol to visualize an increased number of structures that are formed in response to other molecular signals and to further develop the technology to make other areas of the cell accessible.
“This study is only the beginning. Now that we developed this quantitative nanoscale workflow that correlates dynamic signaling behavior with the nano-scale resolution of electron cryo-tomography, we and additional scientists can implement this powerful analytical tool not only for deciphering the inner workings of cell movement but also for elucidating the dynamics of many other macromolecular machines in an unperturbed cellular environment,” says Hanein.
She adds, “Actin is a building-block protein; it interacts with more than 150 actin binding proteins to generate diverse structures, each serving a unique function. We have a surplus of different signals that we would like to map, which could yield even more insights into how cells move.”