Jul 6 2016
Proteins do not work individually but rather form larger complexes similar to the molecular machines that facilitate cell communication, transport cargo in their interiors or replicate their own DNA. Examining and tracking every individual protein inside these machines is critical to the eventually understanding these processes. Nevertheless, super-resolution microscopy that has permitted scientists to visualize closely positioned molecules or molecular complexes with 10-20 nm resolution is not powerful to differentiate individual molecular characteristics inside densely packed complexes.
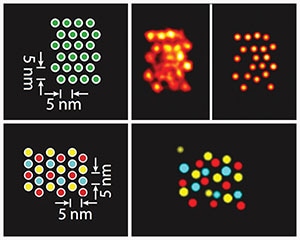 The image shows how the Discrete Molecular Imaging (DMI) technology visualizes densely packed individual targets that are just 5 nanometer apart from each other in DNA origami structures (see schematics on the left). The image on the top right shows a DMI-generated super-resolution image of a clear pattern of individual signals. In the image on the bottom right, three different target species within the same origami structure have been visualized using Exchange-PAINT-enhanced DMI method. (Credit- Wyss Institute at Harvard University)
The image shows how the Discrete Molecular Imaging (DMI) technology visualizes densely packed individual targets that are just 5 nanometer apart from each other in DNA origami structures (see schematics on the left). The image on the top right shows a DMI-generated super-resolution image of a clear pattern of individual signals. In the image on the bottom right, three different target species within the same origami structure have been visualized using Exchange-PAINT-enhanced DMI method. (Credit- Wyss Institute at Harvard University)
A research group headed by Peng Yin, Ph.D., Core Faculty member at Harvard’s Wyss Institute for Biologically Inspired Engineering has, for the first time, been able to distinguish features spaced 5 nm only from each other in a compactly packed, sole molecular structure and to attain the highest resolution until now in optical microscopy. The technology, which is also referred to as "discrete molecular imaging" (DMI), improves the group’s DNA nanotechnology-powered super-resolution microscopy platform with an integrated set of new imaging methods. This study has been published in the July 4th issue of Nature Nanotechnology.
In 2015, the opportunity to facilitate scientists with low-cost super-resolution microscopy using DNA-PAINT-based technologies paved the way for Wyss Institute to commence its spin-off Ultivue Inc.
Yin and his team developed DNA-PAINT technologies, which are based on the momentary binding of two short complementary DNA strands, one of them attached to the molecular target that the scientists plan to envisage and the other one attached to a fluorescent dye. Repetitive cycles of binding and unbinding result in a defined blinking behavior of the dye at the target site. This is highly programmable depending on the choice of DNA strands and has now been exploited further by the researchers’ ongoing work to attain ultra-high resolution imaging.
"The ultra-high resolution of DMI advances the DNA-PAINT platform one step further towards the vision of providing the ultimate view of biology. With this new power of resolution and the ability to focus on individual molecular features, DMI complements current structural biology methods like X-ray crystallography and cryo-electron microscopy. It opens up a way for researchers to study molecular conformations and heterogeneities in single multi-component complexes, and provides an easy, fast and multiplexed method for the structural analysis of many samples in parallel.
Peng Yin, Professor of Systems Biology, Harvard Medical School.
"By further harnessing key aspects underlying the blinking conditions in our DNA-PAINT-based technologies and developing a novel method that compensates for tiny but extremely disruptive movements of the microscope stage that carries the samples, we managed to additionally boost the potential beyond what has been possible so far in super-resolution microscopy," stated Mingjie Dai, study’s first author and a Graduate Student working along with Yin.
The co-authors of the study are Ralf Jungmann, Ph.D., a former Postdoctoral Fellow on Yin’s team and currently a Group Leader at the Max Planck Institute of Biochemistry, Ludwig Maximilian University in Munich, Germany.
The researchers at Wyss Institute have made the ultra-high resolution of DMI using synthetic DNA nanostructures as a benchmark. Subsequently, the scientists propose to apply this technology to real biological complexes like the protein complex that makes a duplicate of DNA in dividing cells or cell surface receptors which bind to their ligands.
"Peng Yin and his team have yet again broken through barriers never before possible by leveraging the power of programmable DNA, not for information storage, but create nanoscale 'molecular instruments' that carry out defined tasks and readout what they analyze. This new advancement to their DNA-powered super-resolution imaging platform is an amazing feat that has the potential to uncover the inner workings of cells at the single molecule level using conventional microscopes that are available in common biology laboratories," said Donald Ingber, M.D., Ph.D., Judah Folkman Professor of Vascular Biology, Harvard Medical School and the Vascular Biology Program at Boston Children’s Hospital, and Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.