Nanoparticle Tracking Analysis, or NTA, determines the particle size distribution of specimens in liquid suspension using the properties of light scattering and Brownian motion. When a laser beam is allowed to pass through the sample chamber, it is scattered by the particles in suspension in its path in such a way that they are easily observable through a 20x magnification microscope equipped with a camera.
The camera runs at roughly 30 fps, recording a video file of the particles under Brownian motion within the field of view of roughly 100 x 80 x 10 µm (Figure 1). The particle movement is recorded on a frame-by-frame basis. The center of each of the observed particles is identified and tracked, and the average distance moved by each particle in the x and y planes is determined by the proprietary NTA software simultaneously.

Figure 1. Schematic of the optical configuration used in NTA.
From this value, the particle diffusion coefficient (Dt) can be determined. Using Dt, the sphere-equivalent hydrodynamic diameter (d) of the particles can be determined using the Stokes-Einstein equation, provided the sample temperature (T) and solvent viscosity (η) are known:
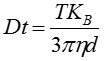
Where, KB = Boltzmann's constant.
NTA is not an ensemble method analyzing a large number of particles, but rather sizing each particle individually irrespective of the others. Figure 2 illustrates the size distribution profile obtained using NTA. Moreover, the movement of the particles is measured within a fixed field of view of roughly 100 x 80 µm, illuminated by a beam of roughly 10 µm in depth.
Figure 2. An example of the size distribution profile generated by NTA. The modal size for this sample is found to be approximately 70 nm, with larger sized particles also present.
These values allow estimating a scattering volume of the sample. A concentration estimation can be achieved in terms of particles per mL for any given size class, or an overall total by determining particle concentration within this field of view, followed by extrapolation to a larger volume.
Viruses and Viral Vaccines
Since NTA allows visualization, sizing, and enumeration of suspension of nanoparticles based on a particle-by-particle basis, it can study virus preparations in higher detail, owing to its ability to identify the concentration and direct number frequency based particle size distribution profile.
In vaccine production, the size of any particular virus or phage particle is less important to estimate virus particle concentration and degree of aggregation. Here, NTA has a significant value due to its ability to measure virus concentration through direct visualization, regardless of virus infectivity.
Virus Characterization, Sizing and Concentration Measurement
Plaque assay, or cell based bioassay in the case of animal cell viruses, typically establishes the titre of phage and virus particles. In these assay systems, the growth of the infective virus particles produces plaques (zones of destroyed cells) that are detectable. It is possible to obtain a direct count of individual infective virus particles, but detecting non-infective virus particles that do not generate plaques is not possible.
Additionally, aggregates consisting of a number of virus particles will generate only single plaques. It is often necessary to determine the number of virus particles in the preparation, whether infective or not, and the degree, if any, to which the preparation is undergoing aggregation. For this purpose, NTA has been evaluated. NTA’s potential for fast and low cost analysis of small volume samples of high polydispersity has been established in previous studies.
Compared to mathematical calculation and spectrophotometer techniques, NTA showed improved accuracy in a study involving the determination of the number concentration of both artificial and natural nanoparticles. However, the potential limitation was the concentration range over which NTA is applicable. Nonetheless, it was demonstrated that NTA could find use in various fields, ranging from vaccine production, drug delivery, and nanoparticle synthesis to aggregation.
NTA has also been evaluated for total virus particle determination, and NTA analysis was employed to measure chromatographic fractions of adenovirus and influenza virus subsequent to purification on a CIM monolithic column. Another study confirmed the ability of NTA to rapidly estimate virus concentration in different samples.
A comparison study of NTA against traditional plaque assay (PA) and quantitative polymerase chain reaction (qPCR) for the detection and analysis of three phage types confirmed the rapid analysis capability of NTA though it is more capital expensive compared to PA and qPCR methods. Moreover, additional reagents were not required. In addition, NTA was able to measure fluorescently labelled virus particles, thereby extending its applicability into harvest materials, for which it is necessary to discern viruses from background host-cell debris and proteins. According to the authors of the study, NTA holds promise for future basic and applied research with bacteriophage.
In another study involving the evaluation of the binding of DNA aptamers to Vaccinia virus and vesicular stomatitis (VSV), researchers of the study concluded that NTA is useful for all further developments of biomolecular binding analyses and aptamer selection. NTA was also used in a study to develop new scalable and reproducible manufacturing processes of adenoviral vectors (human and canine) using stirred tank bioreactors.
Vaccines and Virus-Like Particles
Several research groups have recently reviewed the applicability of NTA in vaccine research. In a study focused on developing large scale production protocols of virus-like particles (VLPs), NTA helped the researchers to confirm that the optimized freestyle culture medium provided nearly double the maximum cell density in batch mode, compared to the use of unsupplemented medium. The researchers also demonstrated that the human embryonic kidney 293 (HEK 293) is the ideal host, owing to several industrially relevant features offered by this cell line, including high transfectability, ability to grow to high cellular densities, ability to grow in suspension, and adaptation to serum-free culture conditions.
In a recent study, NTA technology was used for vaccine characterization and standardization, providing real-time measurement of virus particles in liquid with no particle aggregation or contamination. In another study, researchers proposed new procedures for determining size distribution of biopharmaceuticals by characterizing them using NTA. A research work verified polyplex and lipoplex formations using NTA. In another work, researchers compared the protein content of chaperone-rich cell lysate (CRCL) anti-cancer vaccines prepared from human tumors of different histological origins to assess the homogeneity of their protein content by analyzing the CRCL preparations with NTA and TEM.
Another study evaluated the adjuvant effect of GPI-0100 on a virosomal influenza vaccine formulation using NTA, confirming that very low antigen doses can be used without compromising the protective potential of the vaccine. A research work confirmed that the use of monophosphoryl lipid A (MPLA) can significantly enhance the mucosal and systemic immune responses to pulmonary delivered influenza vaccines by sizing WIV derived from A/PR/8/H1N1 before and after addition of MPLA using NTA. In particular, MPLA-adjuvanted SFD vaccine was very effective, suggesting that adjuvanted vaccine powder to the lungs is a more efficient way of immunizing against influenza.
Phage
In a systematic experimental study on the influence of different bacteriophage (phage) traits on the formation of a plaque, the use of NTA in the determination of phage concentration concluded it necessary to have a more realistic theoretical approach to plaque formation so that the complex interaction between phage and its bacterium host in a spatially restricted environment can be captured. In another study, the high purity and dispersion of the 72 nm fd Y21Mphage preparation used was confimed using NTA.
Virus Templating
NTA allows simultaneous measurement of the average intensity of the scattering of a particle and determination of its size based on its dynamic behavior, thereby enabling distinguishing particles of same size, but with different refractive indices by plotting these two parameters as a function of each other. Using this capability of NTA, virus particles of highly defined structure and uniform size have been employed as templates in the electrodeless deposition metallization process to produce highly monodisperse metallized particles.
A research work showed the use of Cowpea mosaic virus (CPMV) for chemically-coupled-peptide-promoted virus nanoparticle templated mineralization. As anticipated, it was possible to further modify the templated gold nanoparticles with thiol reagents. This research followed previous studies wherein the mineralization of virus was demonstrated using NTA, which has the unique ability to measure both the size and variations in light scattering properties of particles individually.
Many different metals, such as platinum, iron, nickel, cobalt, nickel-iron, cobalt-platinum and gold nanowires have been produced using the plant viral nanoparticle tobacco mosaic virus (TMV), wherein deposition of highly refractive metal layers to the surface of the viruses was observed as a rise in scattered intensity, without a substantial change in particle size, which would have shown aggregation. Virus templated nanoparticles of silica and iron-platinum were prepared in other studies. In one study, the antigenic properties of complexes based on structurally modified plant viruses were analyzed using NTA.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information please visit Malvern Panalytical.