The team hopes this technology will help provide early diagnosis of certain leukemia through detection of gene fusions.
What are MXenes?
The term ‘MXene’ describes a series of two-dimension transition metal carbides, nitrides and carbonitrides. These inorganic compounds were first introduced in 2011 and combine the metallic conductivity of transition metal carbides with a hydrophilic nature.
These materials possess a graphene-like structure that is made by exfoliating MAX phases, novel structural and functional ceramics with a layered structure.
MXene materials have a highly accessible hydrophilic surface compared to graphene and have a large specific surface area, tunable lateral size, as well as good electrical conductivity and mechanical robustness.
These beneficial characteristics make this nanomaterial useful for a range of fields including photocatalysts, supercapacitors, antibacterial agents as well as electrocatalysts for the detection of small molecules.
The latter application would allow for the detection of various molecules from pharmaceutical drugs to environmental pollutants and biomarkers, enabling this to be incorporated into the ideal sensing platform.
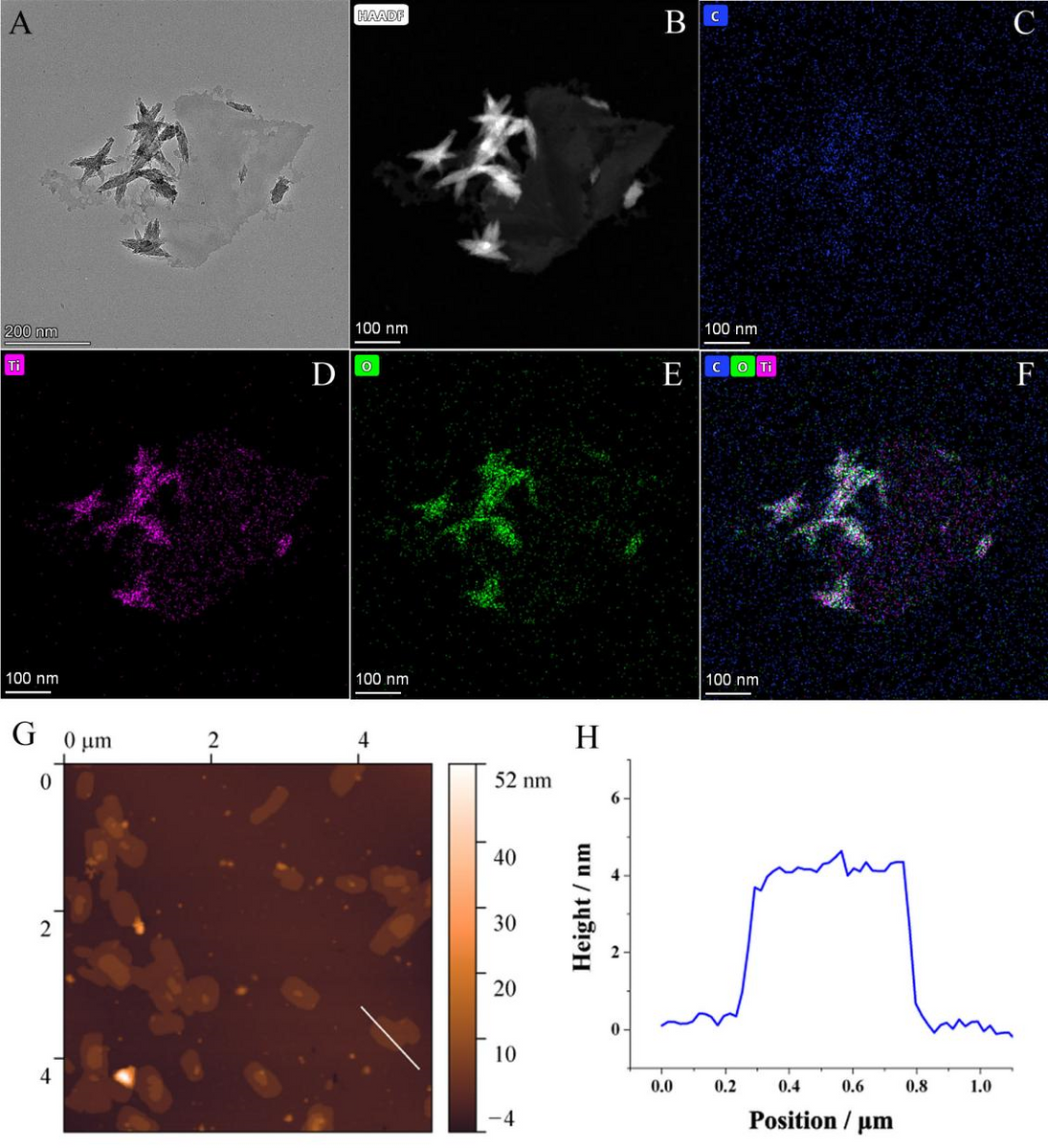
Figure 1. (A) TEM image, (B) HAADF-STEM image and (C-F) STEM-EDS elemental mappings of the Ti3C2Tx MXene flakes. (G) AFM image of the Ti3C2Tx MXene flakes and (H) the height profile along the white line. © Rongjun Yu, Jian Xue, Yang Wang et al (2022)
Necessity for Gene Fusion Detection
Gene fusions consist of the fusion of two previously independent genes which then create hybrid genes that are the result of a structural rearrangement such as translocation.
There have been approximately 10,000 gene fusions identified in human cancers with the first being the BCR-ABL gene fusion, otherwise known as the ‘Philadelphia gene’. This gene fusion was first described in 1960 by Nowell and Hungerford in the city of Philadelphia, with this hybrid gene showing a translocation within chromosomes 9 and 22.
The ABL gene on chromosome 9 was broken and attached to the BCR gene on chromosome 22, creating an abnormally small chromosome 22, with the gene fusion known as BCR-ABL or the Philadelphia gene.
This gene fusion has been found in 95% of all chronic myeloid leukemia (CML) patients as well as being detected in other forms of leukemia.
The detection of gene fusions such as this is a significant component of cancer diagnostics and therapeutics, with cytogenetic testing or PCR tests identifying this genomic alteration.
The sensitivity of gene fusion detection methods as well as for other genomic alterations is critical for the survival of cancer patients. Therein lies the importance of research for this field, with advancements allowing better prognoses for patients through comprehensive detection.
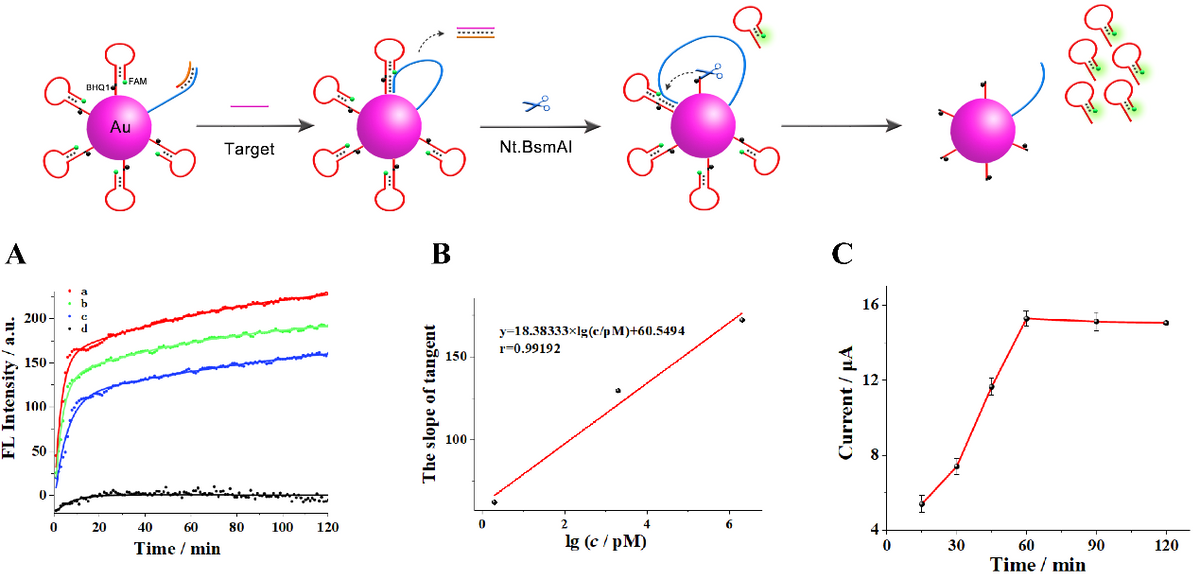
Figure 2. (A) Fluorescence kinetics curves of the nanomachine with addition of 10 U Nt.BsmAI and target BCR/ABL fusion gene at different concentrations (from a to d: 2 μM, 2 nM, 2 pM and blank, separately). (B) The relationship between the slope of tangent at t = 0s and logarithmic value of target BCR/ABL fusion gene concentrations (range of concentration: 2 pM, 2 nM, and 2 uM separately). (C) Time optimization of intermediate DNAs hybridizing with the capture DNA. © Rongjun Yu, Jian Xue, Yang Wang et al (2022)
Advancing Biosensor Application Through Nanotechnology
The researchers of this study have utilized Ti3C2Tx MXene for the advancement of biosensor application, to determine the BCR-ABL gene fusion for clinical diagnosis of CML.
This study consisted of spreading Ti3C2Tx MXene on an electrode along with gold nanoparticles which functioned to aid with DNA capture probe immobilization.
The electrocatalytic activity of Ti3C2Tx MXene for phenols oxidation assisted the researchers in constructing a catalytic amplification strategy for electrochemical biosensors to detect the BCR-ABL biomarker.
Ultimately, the generation of a DNA walking machine that would expose DNA fragments in the presence of this particular gene fusion, and cascading catalysis for signal amplification, which was amplified through the Ti3C2Tx MXene-catalytic electrochemical oxidization, enabled the biosensor to achieve significant results.
The sensitivity of detection of BCR-ABL was proven to be highly effective through this innovative strategy, enabling this to be a powerful tool for bioanalysis and diagnosis of CML.
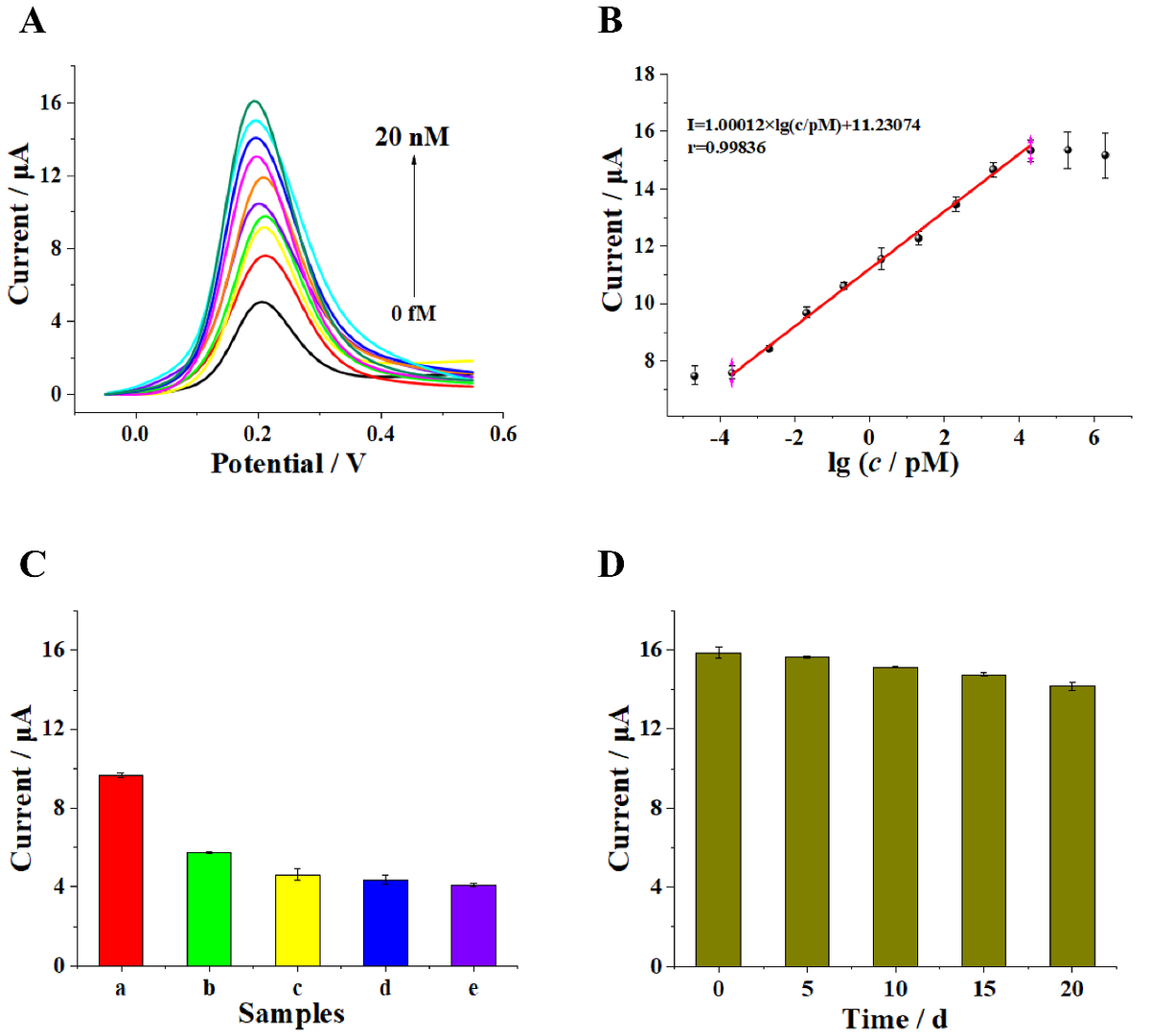
Figure 3. Evaluation of the sensitivity and specificity of the biosensor: (A) DPV curves response of the electrochemical biosensor upon the increase in target BCR/ABL fusion gene concentration (from bottom to top: 0 fM, 0.2 fM, 2 fM, 20 fM, 200 fM, 2 pM, 20 pM, 200 pM, and 2 nM, 20 nM, separately) and (B) the corresponding linear relationship between DPV signal and logarithmic value of target BCR/ABL fusion gene concentrations (range of concentration: 20 aM, 0.2 fM, 2 fM, 20 fM, 200 fM, 2 pM, 20 pM, 200 pM, and 2 nM, 20 nM, 200 nM, 2 uM separately). (C) DPV responses of the electrochemical biosensor to different oligonucleotides (20 fM): (a) BCR/ABL fusion gene (target), (b) single-base-mismatched strand (B1), (c) two-base-mismatched strand (B2), (d) noncomplementary strand (B3), and (e) blank. (D) Stability of the proposed biosensor. The error bars represent the standard deviation of three parallel measurements. © Rongjun Yu, Jian Xue, Yang Wang et al (2022)
Future Outlook
This research illustrates the great potential of Ti3C2Tx MXene nanozyme due to its efficient electrocatalysis activity for phenols oxidation, both for the field of biosensors with successful detection of BCR-ABL gene fusions as well as potentially for other fields, including sewage treatment and organic synthesis.
The field of biomedical research and cancer diagnostics, as well as therapeutics, is continually advancing and with innovative nanotechnology research, the development of novel ultrasensitive detection tools can aid with reducing the burden of global diseases.
The American Cancer Society has estimated 8,860 cases of CML will emerge in 2022 in the United States, with 1,220 people dying from this disease; the translation of powerful bioanalysis tools can assist with decreasing this number with earlier detections which can lead to earlier treatment through the use of a tyrosine kinase inhibitor.
This research can also be a stepping-stone for advancing the detection of other gene fusions such as the NTRK gene fusion, which is the oncogenic driver for non-small cell lung cancer.
The detection of small molecules through this strategy can be useful in many health-related diseases which will ultimately transform conventional medicine towards a more personalized approach.
*Important Notice
Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information
Reference
Rongjun Yu, Jian Xue, Yang Wang et al Novel Ti3C2Tx MXene Nanozyme with Manageable Catalytic Activity and Application to Electrochemical Biosensor, 15 February 2022, PREPRINT Available at: https://doi.org/10.21203/rs.3.rs-965061/v2, https://www.researchsquare.com/article/rs-965061/v2
Further Reading
Bruno, R. and Fontanini, G., (2020) Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics, 10(8), p.521. Available at: 10.3390/diagnostics10080521
Cancer.org. (2022) Key Statistics for Chronic Myeloid Leukemia. [online] Available at: https://www.cancer.org/cancer/chronic-myeloid-leukemia/about/statistics.html
Haider MZ, Anwer F. Genetics, Philadelphia Chromosome. [Updated 2021 Jul 21]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK560689/
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.