May 24 2018
It is paramount to understand a material’s microscopic structure in order to understand how it works and its functional properties. Developments in fields like materials science have progressively boosted abilities to establish these features to even higher resolutions. One method for imaging at nanoscale resolution, transmission electron microscopy (TEM), is one instance of a promising technology in this field. Researchers have found a new way to harness the power of TEM to measure a material’s structure at the highest possible resolution - defining the 3D position of each individual atom.
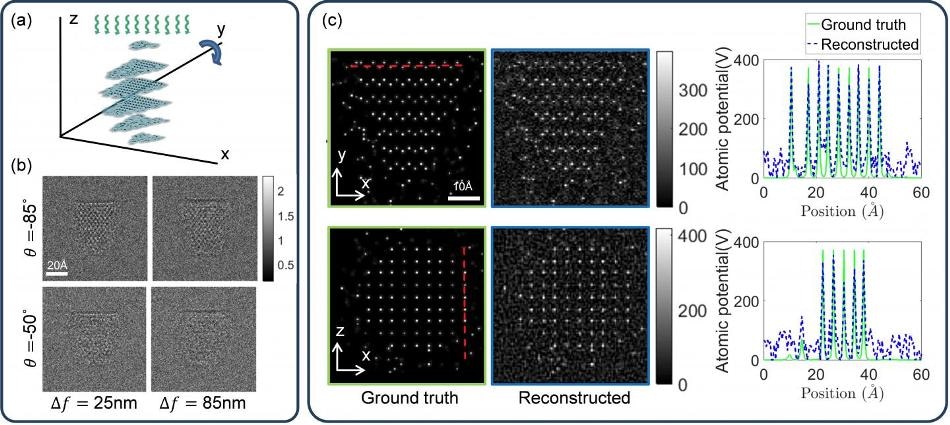 Schematic of proposed TEM 3D atomic imaging with multi-slice method with four examples of noisy intensity measurements at different angles of rotation, and 3D atomic potential reconstructions and 1D cross-sections along x and y directions. (Image Credit: David Ren)
Schematic of proposed TEM 3D atomic imaging with multi-slice method with four examples of noisy intensity measurements at different angles of rotation, and 3D atomic potential reconstructions and 1D cross-sections along x and y directions. (Image Credit: David Ren)
Presenting their study at the OSA Imaging and Applied Optics Congress, held between 25 and 28 June, in Orlando, Florida, USA, a team of scientists have proved a method using TEM tomography to establish the 3D positions of strongly scattering atoms. Through imitation, the team demonstrated that reconstruction of the atomic potentials with atomic resolution using just image intensity measurements is possible, and that it’s possible to achieve that on molecules that are highly sensitive to electron beams.
Transmission electron microscopy is used extensively in both materials science and biology. Because we fully solve the nonlinear propagation of the electron beam, our tomographic reconstruction method will enable more quantitative reconstruction of weakly scattering samples, at higher or even atomic resolution.
Colin Ophus - National Center for Electron Microscopy, Lawrence Berkeley National Lab & Member of the Research Team
Just like the way computerized tomography (CT) scans for medical imaging in hospitals are built using a series of 2D cross-sectional images at varying increments, electron tomography builds a 3D volume by rotating samples incrementally, gathering 2D images. While most CT imaging in hospitals is performed using X-rays to establish features of larger things like bones, the beams of electrons used in TEM allows scientists to observe with considerably higher resolution, down to the atomic scale.
However, on the atomic scale we cannot neglect the very complex quantum mechanical effects of the sample on the electron beam. This means in our work, we must use a much more sophisticated algorithm to recover the atomic structure than those used in an MRI or CT scan.
Colin Ophus - National Center for Electron Microscopy, Lawrence Berkeley National Lab & Member of the Research Team
The TEM configuration the team used measures the energy intensity that reaches the microscope’s sensor, which is proportional to the number of electrons that reach the sensor, a number that relies on how the electron beam is arranged for each experiment. Using the intensity data, the new algorithm formulated by the team stitched the 2D projected images into a 3D volume.
Computers will be taxed exponentially more than handling single 2D images if there is a jump to three dimensions with large fields of view. To sort this, they altered their algorithm to be used on graphic processing units (GPUs), which can perform several times more mathematical operations in parallel than standard computer processing units (CPUs).
“We are able to obtain results in a reasonable amount of time for realistic sample dimensions,” said David Ren, a member of the team.
With normally weaker bonds between their atoms, biomolecules can be infamously hard to examine using TEM because the electron beams used to investigate a metal alloy, for instance, would usually tear a biomolecule apart. Decreasing the electron dosage in a sample, though, can form images that are very noisy; other algorithms presently in use cannot reconstruct a 3D image. Owing to a more precise physical model, the team’s new algorithm has the capability.
Now that they have completely established the reconstruction algorithm, the team said they want to apply what they’ve witnessed from simulations to experimental data. They plan to share all of their reconstruction codes as open source for the research community.