Jan 25 2016
Scientists have successfully measured the dynamical behavior of the organic ligand layer of gold nanoparticles in water. This provides a better understanding of the structure of nanometer-scale gold particles in solution, and will allow the creation of controllable strategies for the functionalization of ligated nano-sized particles for a variety of applications. Researchers from the Colorado State University, USA and the University of Jyväskylä, Finland were involved in the study, and the results have been published in Nature Communications.
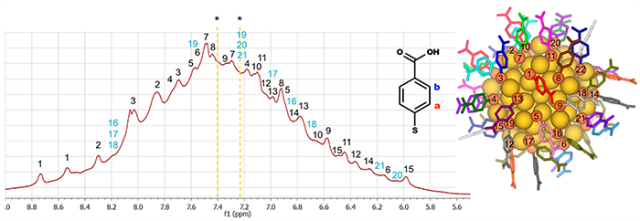 The proton NMR spectrum originating from the ligand layer of the Au102 nanoparticle in water (left). The spectrum has been fully interpreted by assigning the observed signals (peaks) to all of the 22 symmetry-unique thiol ligands numbered in the solid state structure of the Au102 particle (right).
The proton NMR spectrum originating from the ligand layer of the Au102 nanoparticle in water (left). The spectrum has been fully interpreted by assigning the observed signals (peaks) to all of the 22 symmetry-unique thiol ligands numbered in the solid state structure of the Au102 particle (right).
Gold nanoparticles are often studied for use as components in molecular and photonics electronics, drug delivery devices, sensors, catalysts, and biological contrast agents. The smallest nanoparticles feature metal cores of just 1 nm to 2 nm, with a few tens to a couple of hundred gold atoms. A stabilizing organic ligand layer encloses these metal cores. Most of these compounds are known as "clusters", whose solid-state atomic structure and molecular formulas have been determined over the past few years. Despite this, it is still difficult to interpret their dynamical behavior and atomic-scale structure in the solution phase. This data is critical and can aid scientists to gain a better understanding of the interaction between the nanoclusters and the environment.
The scientists examined a molecularly accurate nanocluster, which contains 44 thiol ligands and 102 gold atoms. In 2007 single-crystal X-ray diffraction experiments were used to determine the cluster’s solid-state structure. Featuring a low symmetry, the ligand shell creates signals in conventional proton-nuclear magnetic resonance (NMR) measurement. Through a combination of associated NMR experiments, molecular dynamics simulations, and density functional theory calculations, the team were able to acquire a complete assignment of the entire signals to particular thiol ligands. This specific cluster material was previously used by scientists at Jyväskylä for structural analysis of enteroviruses.
Now that we know exactly which ligand produces which NMR signal, we can proceed with precise studies on how this nanocluster interacts with the chemical and biological environment in the water phase. This gives unprecedented potential to understand and control the inorganic-organic interfaces that are relevant to hybrid inorganic-biological materials
Hannu Häkkinen, Academy Professor, Nanoscience Center, University of Jyväskylä.
The study, performed by the Finnish and US researchers, were coordinated by Häkkinen.
Other scientists who were involved in the study are Sami Malola, Kirsi Salorinne, Hannu Häkkinen, and Xi Chen from the University of Jyväskylä; and Christopher J. Ackerson, Christopher D. Rithner, and O. Andrea Wong from Colorado State University.
The computational work was carried out at the CSC – the Finnish IT Centre for Science. The Academy of Finland supported the study.