Nov 29 2018
Motions of protein molecular motors can be observed using optical microscopy while they are at work in the biological systems. It would be possible to track the motions of individual molecules by labeling molecules of interest with suitable probes.
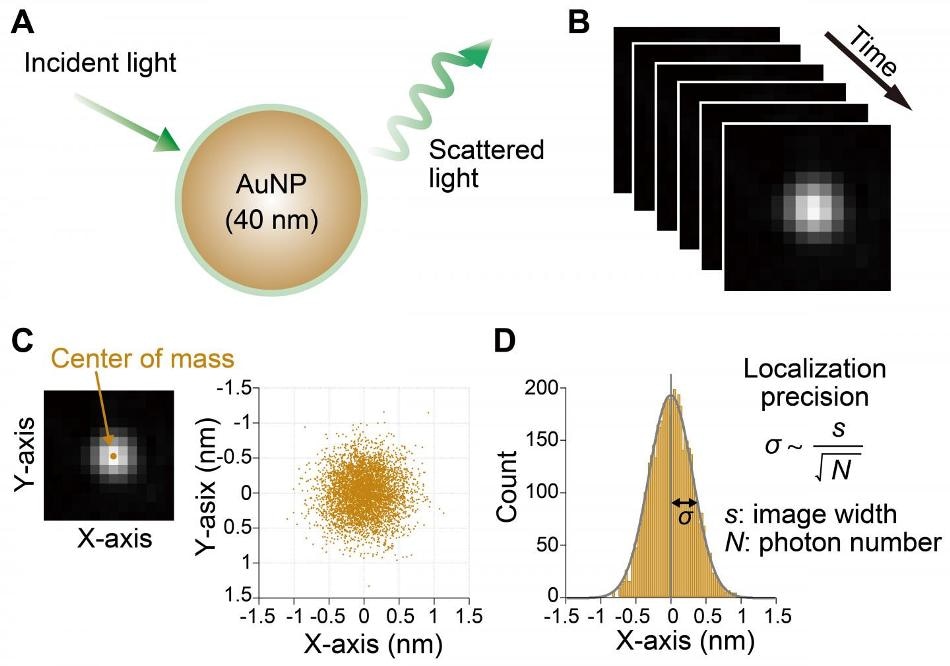 Flowchart to estimate localization precision of AuNPs. (A) Schematic of 40 nm AuNP scattering incident light. (B) Sequential acquisition of scattering images of AuNP. (C) Two-dimensional plot of time course of the center position. (D) Distribution of center position of AuNP along X-axis. Standard deviation of distribution is defined as the localization precision. (Image credit: NINS/IMS)
Flowchart to estimate localization precision of AuNPs. (A) Schematic of 40 nm AuNP scattering incident light. (B) Sequential acquisition of scattering images of AuNP. (C) Two-dimensional plot of time course of the center position. (D) Distribution of center position of AuNP along X-axis. Standard deviation of distribution is defined as the localization precision. (Image credit: NINS/IMS)
Localization precision (the precision with which the position can be determined) of a moving molecule is mainly determined using the number of photons captured by a single image frame. Fluorescent probes can be used for achieving a localization precision of 1 nm and millisecond time resolution, by capturing around 10,000 photons. Further enhancement of the localization precision is critical for gaining deeper insights into the operation mechanisms of molecular motors, but the lesser number of photons acquired from the fluorescent probe has limited the improvement. Recently, gold nanoparticles (AuNPs) with the ability to strongly scatter the incident light have been used as an alternative for the fluorescent probes. In earlier research works, microsecond time resolution has been realized using AuNPs. Yet, the basic limit of the localization precision has not been experimentally tested in-depth.
In this research, Ando and colleagues from the Institute of Molecular Science, Japan, used newly developed annular illumination total internal reflection dark-field microscopy to successfully achieve atomic-level, 1.3 angstroms (Å) localization precision with 40 nm AuNPs at a time resolution of 1 ms. Moreover, 5.4 Å localization precision has been successfully realized even at a time resolution of 33 μs.
First, the researchers investigated the fundamental law that restricts the localization precision of AuNP and validated that the localization precision is enhanced in proportion to the square root of the photon number, similar to earlier theoretical considerations. Moreover, the lower limit of the localization precision using a dark-field imaging system developed earlier by the researchers was about 3 Å, which was limited by signal saturation of the detector. They developed a new annular illumination total internal reflection dark-field microscopy to further enhance the localization precision. This system shapes the laser light into a ring to be able to use higher laser intensity than the earlier system without any damage to the objective lens. In addition, signal saturation was suppressed by applying smaller pixel size of the detector (larger number of pixels in an image). These enhancements allow a localization precision of 1.3 Å to be achieved at a time resolution of 1 ms.
Moreover, the research performed a detailed analysis of the link between localization precision and time resolution. The newly developed system realized a localization precision of 5.4 Å even at a time resolution of 33 μs. The researchers also analyzed the influence of size on the localization precision to reduce possible steric hindrances of AuNPs on the protein molecular motors, thus achieving a localization precision of 1.9 Å at a time resolution of 1 ms with 30 nm AuNPs.
Subsequently, they used the newly developed imaging system to observe in detail the stepping motions of a dimeric linear molecular motor kinesin, moving along the microtubule (Fig. 3). One motor domain (head) of the kinesin was labeled by 40 nm AuNP, and the motion was captured at a time resolution of 10 μs. In an earlier research, unbound head exhibited diffusional motion only at the right side of bound head on microtubule. This outcome indicates the unidirectional rotation of two heads of kinesin during linear motion. High-speed, high-localization single-particle tracking carried out as part of the research successfully unraveled details of the transition from bound to unbound states of the kinesin head. Since there were no evident leftward trails observed, the researchers concluded that the kinesin in fact rotates unidirectionally during the linear motion.
Apart from kinesin, the methodology developed in this research can be applied to capture the dynamics of different protein molecular motors with atomic-level localization precision and microsecond time resolution. As a matter of fact, recently, the researchers resolved the forward and backward 1-nm steps during fast unidirectional motion of a chitinase driven by processive catalysis, and it was found that processive chitinase functions as a “burnt-bridge” Brownian motor. Consequently, the newly developed system will essentially contribute to gain further insights into the operation mechanisms of various protein molecular motors. Moreover, it is also possible to use the system to visualize atomic-level motions of synthetic molecular motors, which are considerably smaller than the protein molecular motors.