Feb 15 2019
Researchers at Rice University have expanded the capabilities of conventional laboratory microscopes by adding a new dimension to their revolutionary technique.
 Rice University researchers have created a method to design custom masks that transform 2D fluorescent microscopy images into 3D movies. (Image credit: the Landes Research Group)
Rice University researchers have created a method to design custom masks that transform 2D fluorescent microscopy images into 3D movies. (Image credit: the Landes Research Group)
A couple of years back, super-temporal resolution microscopy was introduced by the Rice lab of chemist Christy Landes. Through this technique, scientists were able to image fluorescent molecules in a much faster way—as much as 20 times faster than normally allowed by conventional lab cameras. Now, they have created a standard technique to allow a microscope to capture 3D spatial data together with molecular movement—the fourth dimension—over time.
According to the researchers, this technique will assist scientists focused on dynamic processes to view the location of target molecules and also the speed at which they move—for instance, inside living cells. The novel technique developed by the Rice team widens the capabilities of prevailing wide-field fluorescence microscopes. The results of the study have been reported in the team’s open-access paper in Optics Express.
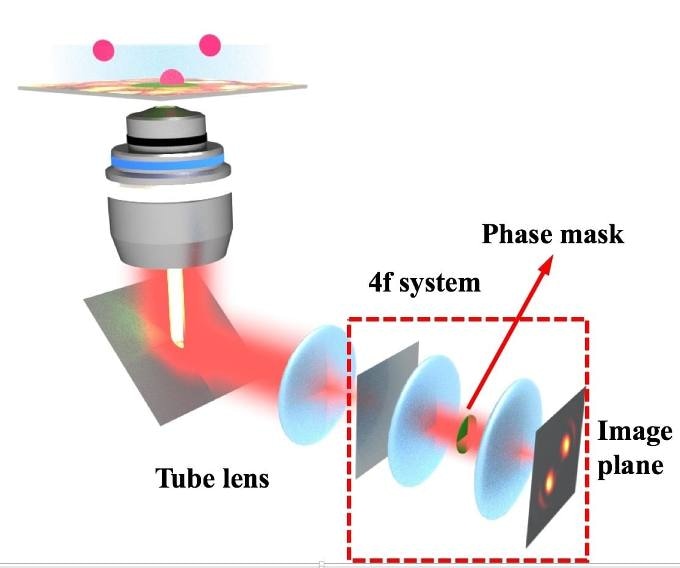
It details the development of custom phase masks—spinning and transparent disks that exploit the light’s phase to alter the shape of the image taken by the camera of the microscope. The shape includes data related to the 3D position of a molecule in space and how it behaves within the camera’s field of view over time.
The blurry blob—which seems to be an inconvenience—in a microscope image is converted into an asset by a phase mask. Investigators have dubbed this blob as point spread function and apply it to obtain information related to objects below the diffraction limit that are relatively smaller than all visible light that can be viewed by microscopes.
In the original study, a rotating phase mask was applied that converted light from one fluorescent molecule into the so-called rotating double helix, termed by the researchers. The image that was captured appeared as two glowing disks on the camera, similar to the globes of a barbell. In the latest study, the rotating barbells allowed the researchers to see where exactly the molecules were in 3D (three-dimensional) space and, at the same time, provided a time stamp to each molecule.
The core of the latest study lies in algorithms developed by Wenxiao Wang, Rice electrical and computer engineering alumnus and lead author. The algorithms make it feasible to create custom phase masks that alter the point spread function’s shape.
“With the double helix phase mask, the time information and spatial information were connected,” stated Chayan Dutta, co-author and a postdoctoral researcher in Landes’ laboratory. “The lobes’ rotation could express either 3D space or fast time information, and there was no way to tell the difference between time and space.”
That problem can be solved by better phase masks, he stated. “The new phase mask design, which we call a stretching lobe phase mask, decouples space and time,” said Dutta. “When the targets are at different depths, the lobes stretch farther apart or come closer, and the time information is now encoded just in the rotation.”
The trick is to improve the pattern for varied depths by manipulating light at the spinning phase mask, and this is achieved by the refractive pattern, which is programmed within the mask through the algorithm.
Each layer is optimized in the algorithm for different detection depths. Where before, we could see objects in two dimensions over time, now we can see all three spatial dimensions and fast time behavior simultaneously.
Nicholas Moringo, Study Co-Author and Graduate Student, Department of Chemistry, Rice University.
Wide-field fluorescence microscopes are used in many fields, especially cell biology and medical imaging. We are just starting to demonstrate how manipulating light’s phase within a microscope is a reasonably simple way to improve space and time resolution compared to developing new fluorescent tags or engineering new hardware improvements.
Christy Landes, Professor, Chemistry and Electrical & Computer Engineering, Rice University.
She said that one major outcome that could have wide impact is that the investigators generalized the design of the phase mask, which means scientists can develop masks to create almost any random pattern. In order to prove this, the researchers designed and developed a mask to create an intricate point spread function that clearly spells out “RICE” at varied focal depths.
The below video shows that as the microscope moves to different depths below and above the focal plane, the ghostly letters appear and disappear. Flexibility like this will be valuable for applications, for instance, examining processes within living cancer cells—a project the lab is hoping to follow up soon with partners at Texas Medical Center.
If you have a cell on a glass slide, you’ll be able to understand where objects in the cell are in relationship to each other and how fast they move. Cameras aren’t fast enough to capture all of what happens in a cell, but our system can.
Nicholas Moringo, Study Co-Author and Graduate Student, Department of Chemistry, Rice University.
Study co-authors are Rice graduate student Fan Ye; former Rice postdoctoral researcher Hao Shen, currently an assistant professor at Kent State University; and Jacob Robinson, a Rice assistant professor of electrical and computer engineering. Landes is a professor of chemistry and of electrical and computer engineering. Wang is currently a software engineer at Google.
The study was supported by the National Science Foundation and the Welch Foundation.
RICE phase mask
Rice University researchers have created an algorithm to produce custom phase masks that help analyze molecular processes below the diffraction limit and 20 times faster than traditional cameras. The team produced a mask that shows the depth of an object by spelling out RICE as the focal plane changes. (Video credit: Wenxiao Wang/Landes Research Group)