Quantifying the protein interaction kinetics is critical for biosensor development, disease discovery biomarkers, cognizance of biological mechanisms, and drug candidate identification. Label-free methods like surface plasmon resonance (SPR) require the introduction of binding ligands to functionalized sensor surfaces installed with microfluidic systems that are difficult to maintain. Moreover, this process involves immobilizing bound proteins to the sensor surface using conjugation chemistry and blocking.
Acquisition of purified proteins is via affinity separation like magnetic nanoparticles. In this process, proteins are trapped by nanoparticles which are surface modified by affinity probes. It is a tedious process involving the elution of proteins and buffer exchange. Hence the protein purification process needs to be simplified.
Measuring the size of molecule-bound nanoparticles is challenging due to their small size. Novel methods like charged coupled device (CCD) detectors and nanoparticle tracking analysis (NTA) has outperformed the dynamic light scattering (DLS) method in analyzing the variation in size of single nanoparticles.
Novel Method for Protein Purification and Analysis
In the present work, the authors introduced a new method based on NTA to quantify protein binding kinetics for the proteins trapped on the magnetic nanoparticles.
The advantage of this method is that it can eliminate the need for protein elution from the nanoparticles or immobilization on the sensing surface. The change in nanoparticle size after ligand binding was tracked optically. The current approach is a novel method that makes protein purification and subsequent analysis an easy process.
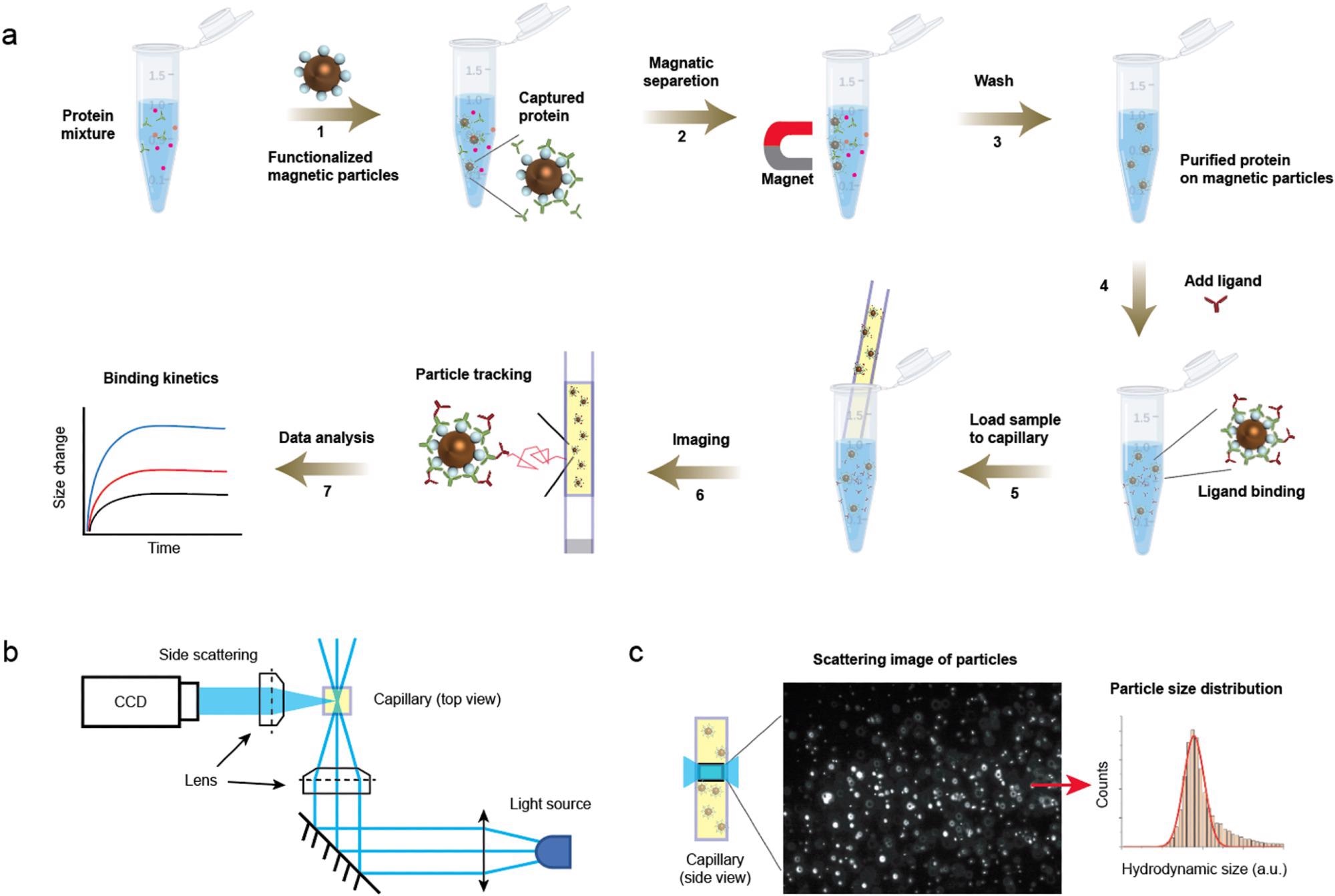
Detection principle. (a) Protocols showing capture of protein to the magnetic particles (steps 1–3) and the subsequent ligand binding detection (steps 4–7). After adding ligand (step 4), a small volume of the mixture was immediately transferred to a capillary, which was mounted on the imaging setup for particle motion tracking. (b) Optical setup for particle tracking. The incident light is focused on the capillary and side scattered light of particles are imaged. (c) A zoom-in of the capillary showing the region being imaged (left) and an image showing the scattering light of the particles (middle). The particles are tracked, and the hydrodynamic radius is extracted from thousands of particles (right). © Zhao, Y., Ma, G., & Wang, S. (2022).
Research Findings
The single measurement to determine particle size integrates a broad distribution that influences the precision and accuracy. The distribution is fitted into a Gaussian equation with standard deviation σ and mean value r. Furthermore, for each measurement, the values of σ and r were different.
The authors discussed the peak broadening and measurement variation possibilities. Since reducing the dimension does not lose information for Brownian motion, they performed a Monte Carlo simulation for the unidimensional model.
In the simulation, the authors studied four parameters distribution of particle size, tracking duration distribution, number of tracks from each particle, and particle number. The studies began with a simple case, and the increased complexity in scenarios mimics the experimental conditions.
Results revealed that the particle number and track duration greatly influence precision and accuracy. Adequate tracks reduced the variance and improved the precision, and the extended tracking time reduced the fitting error and improved accuracy.
The authors considered false track as a second noise source, obtained due to impurities and aggregates, and influenced the fitting quality. Moreover, false tracks generate inaccurate size information due to short track information and blinking image intensities. Laser heating induced flow was the third noise source, which led to particle drifting.
The mean square displacement (MSD) contained Brownian motion and drift information, and drift increases the MSD value and reduces the hydrodynamic size. Likewise, increasing the temperature increases MSD.
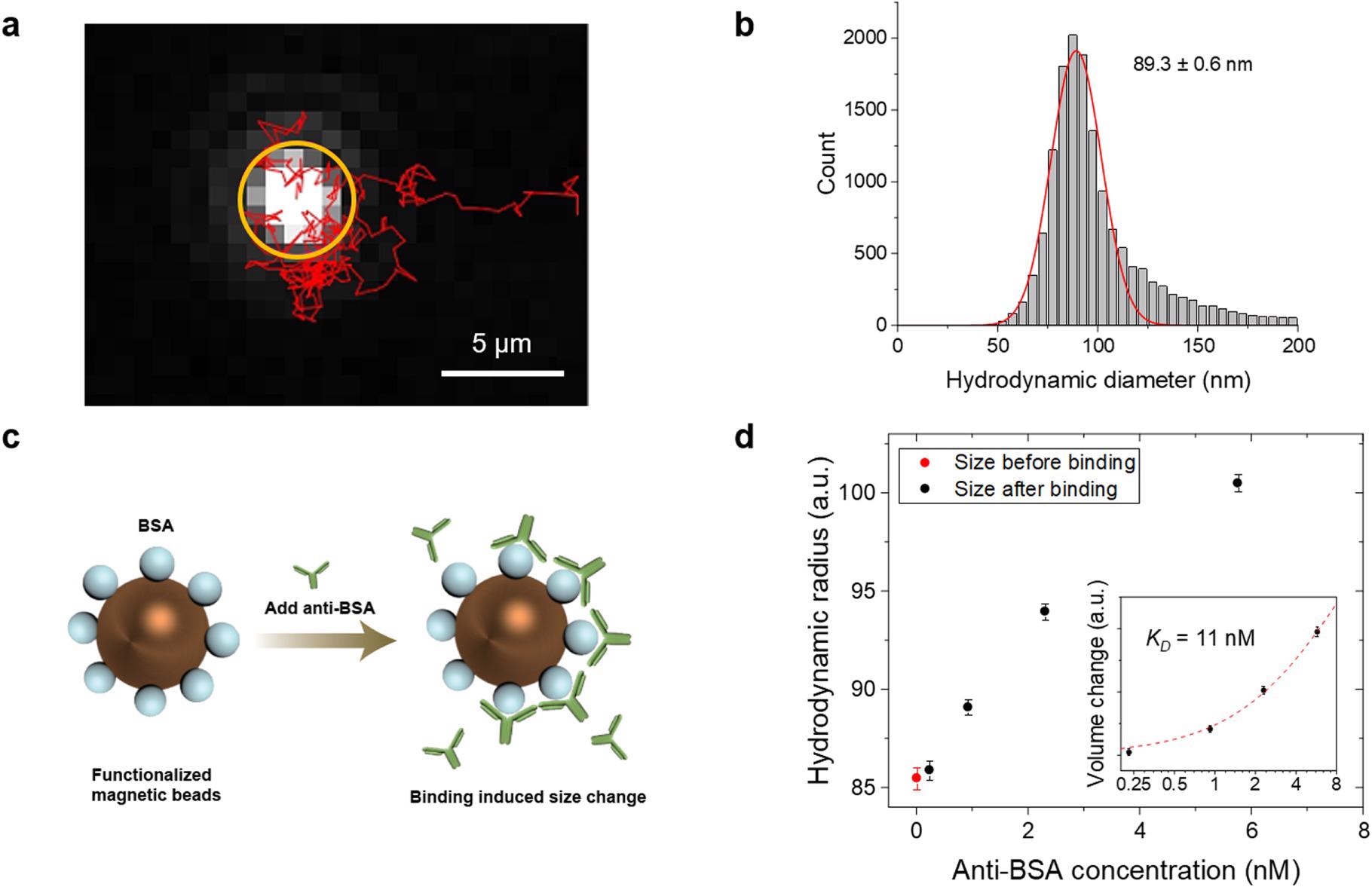
Measuring particle size and ligand binding affinity. (a) Tracking the motion of a single particle. The bright spot in the orange circle is the particle, and its motion within 5.2 s is tracked (red trace). (b) Histogram showing the diameter distribution obtained from the track of >2000 particles. (c) Anti-BSA is added to BSA coated particles, and the binding increases the particle size. (d) Hydrodynamic diameter of the particles as a function of anti-BSA concentration at equilibrium. The inset shows plotting the data in log-log scale, where the red dotted curve is fitting of the data to Eq. 2. The error bars represent standard deviation of >2000 individual particles. © Zhao, Y., Ma, G., & Wang, S. (2022).
The present method can quantify the antibody binding kinetics at concentrations that cover the range in immunoassays. While the upper limit of concentration is due to particle concentration because of false tracking, the lower limit is due to the size of the molecule and binding affinity. To measure the binding kinetics and dynamic range, particles were mixed with the ligand and loaded into a capillary from a centrifuge tube. The mixing of samples and settling down after loading took approximately two minutes. Subsequent calculations allowed the authors to investigate the possibilities for the dynamic range broadening.
Furthermore, reducing the preparation time from two minutes to one second increased the measurable range by two-fold. The decrease in σ to 0.1% extended the measurable range two-fold, covering all the molecular interactions. Here the microfluidic device reduced preparation time, which provided fast mixing and controlled flow.
Increasing the particle count number lowered the noise. Although noise was a statistical issue, tracking the particles for an extended period overcame this issue.
Conclusion
In the present work, the authors quantified the protein-protein interaction by employing a novel particle tracking method. This approach allows the pulling down of proteins for subsequent separation using magnetic nanoparticles. Moreover, the measurement of protein binding kinetics did not involve elution or immobilization.
The authors anticipate that the present method can streamline the protein purification process and analysis that expedites protein research.
Reference
Zhao, Y., Ma, G., & Wang, S. (2022). Magnetic Nanoparticle Tracking for One-Step Protein Separation and Binding Kinetics Analysis. Journal of The Electrochemical Society. https://iopscience.iop.org/article/10.1149/1945-7111/ac6bc5
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.