Jul 27 2009
Tracking down new active agents for cancer or malaria treatment could soon become easier - thanks to a computer program with which researchers from the Max Planck Institute of Molecular Physiology in Dortmund aim to facilitate the search for suitable pharmaceutical substances. The program, which is called Scaffold Hunter, acts as a tool for navigating chemical space. It generates maps of chemically-related structures and links them to biological activity, that is, to their potential to bind to proteins, in particular medically relevant proteins. With the help of this new tool, the Max Planck scientists, together with colleagues from the universities of Frankfurt, Eindhoven and New Mexico, have identified substances that could provide possible candidates for the development of active agents for use in cancer treatment and malaria. (Nature Chemical Biology, June 28, 2009).
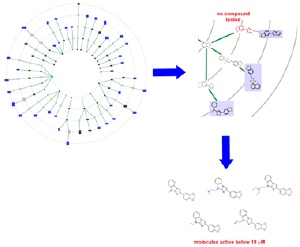 The search for active agents in the tree of structures: Basic chemical scaffolds are linked initially on the basis of their structural similarity. Compounds that influence the enzyme pyruvate kinase are shown in blue, the virtual scaffolds in grey. Variants of the virtual scaffold shaded in red (top right) were tested for their biological potency and pointed to other active substances (bottom right)). Image: MPI of Molecular Physiology
The search for active agents in the tree of structures: Basic chemical scaffolds are linked initially on the basis of their structural similarity. Compounds that influence the enzyme pyruvate kinase are shown in blue, the virtual scaffolds in grey. Variants of the virtual scaffold shaded in red (top right) were tested for their biological potency and pointed to other active substances (bottom right)). Image: MPI of Molecular Physiology
The dimensions of the chemical space, which contains the total number of all conceivable chemical structures, are unimaginable: it is estimated to contain up to 10160 different molecules. Written out in full, this figure would fill two lines of closely-spaced numbers on a typed page. However, only some of these - 1060 molecules according to the estimates - are potential active agents. Identifying these islands of biological activity in the ocean of all potential compounds is not an easy task.
"Organic synthesis cannot gauge the chemical space in its entirety," explains Stefan Wetzel, a researcher from Herbert Waldmann’s group at the Max Planck Institute of Molecular Physiology in Dortmund. Chemists cannot cook up all possible compounds to test them. Therefore, the researchers have developed this navigation system to help them steer their way through this sea of possibilities. The program Scaffold Hunter generates a map of the chemical space based on structural criteria and uses it to identify biologically active compounds, e.g. natural substances. The program can also be used to predict new candidate agents that do not occur in nature.
The scientists focus on the medically relevant section of the chemical space, in which molecules contain ring-shaped structures. To do this, they reduce the molecules to their characteristic scaffolds. Scaffold Hunter then orders these structures in a kind of family tree based on their similarities: the program assigns smaller ‘parent’ scaffolds to each scaffold by gradually removing rings from the original ‘child’ scaffold. This generates innumerable parent-child relationships - structurally related molecules of varying complexity. The advantage lies in the fact that chemically similar compounds are very likely to display similar biological activity.
"These structurally-based lineages form the branches of the tree," explains Stefan Wetzel: "With the help of Scaffold Hunter we move along the branches from complex to increasingly simple structures which may be similar in their effect." Thus, the researchers identify structurally simple scaffolds with promising characteristics as the starting point in the quest for new active agents: chemists can then vary the scaffolds with different appendages to synthesize the optimal active agent. Scaffold Hunter can also be used to predict bioactive molecules that do not arise in nature but are very likely to display similar activity to related natural molecules, as the program also creates and visualizes virtual scaffolds. The researchers immediately demonstrated how efficiently the program works by discovering new inhibitors of pyruvate kinase. The inhibition of this enzyme is seen as a promising approach to the treatment of cancer and malaria.
An even more detailed search can be carried out if the scientist can enter information about biological activity - if available - at the beginning of the navigation process. In this case, Scaffold Hunter only links the scaffolds that are known to display the same biological activity to the branches. As a result, these branches are very likely to bear fruit: active substances are probably also located in the branches between the substances whose biological activity is already known. "In this way, we tracked down new inhibitors for 2-lipoxygenase and the oestrogen receptor alpha," says Steffen Renner, a former researcher at the Max Planck Institute and now an employee of the Novartis pharmaceutical concern. 5-lipoxygenase is a target protein in the treatment of inflammation and bladder cancer, while the oestrogen receptor alpha is an important starting point in the treatment of breast cancer.
"Scaffold Hunter is a key technological tool with innumerable possible applications," says Stefan Wetzel. "The program was consciously designed in a very user-friendly way so that non-experts can use it to analyse their data themselves," he adds. The researchers have made Scaffold Hunter available free of charge on the Internet. The source code can also be obtained: advanced users can thus adapt the program to their requirements and embark on more targeted explorations of the chemical space.