Dec 2 2009
An important Brandeis study appearing in the December 3 issue of Nature raises the curtain on the hidden lives of proteins at the atomic level. The study reports that for the first time, researchers used x-ray crystallography and nuclear magnetic resonance (NMR) techniques to directly visualize protein structures essential for catalysis at the rare high-energy state. The study also showed how the motions of these rare, or hidden, structures collectively, directly contribute to enzyme catalysis.
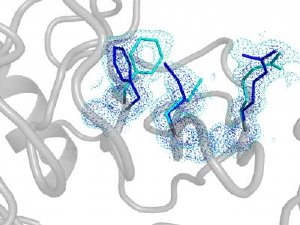 The hidden alternate structure of the human prolyl isomerase uncovered by ambient temperature high-resolution x-ray crystallography. Fast flipping between the main (blue) and minor (light blue) structures is essential for catalytic power. Credit: Dorothee Kern
The hidden alternate structure of the human prolyl isomerase uncovered by ambient temperature high-resolution x-ray crystallography. Fast flipping between the main (blue) and minor (light blue) structures is essential for catalytic power. Credit: Dorothee Kern
In doing so, the study also suggests new molecular sites for potential drug targets, the cornerstone of rational drug design. Drugs may bind, or dock, to the infrequent high-energy states of target enzymes that have been hidden to traditional structural methods. The thinking is that drugs can be designed by docking algorithms to a collection of protein structures, not just one, providing better bio-molecular targeting.
This study comes in the wake of earlier Brandeis studies aimed at advancing understanding of protein function using pioneering techniques such as NMR. For a long time, scientists viewed proteins more or less as macromolecular wallflowers, venturing out onto the atomic-level dance floor to perform only during catalysis, their active state.
Then, several years ago, Brandeis biophysicist Dorothee Kern reported in Nature that her lab's experiments using NMR also linked protein function to their much rarer high-energy state, in the absence of catalysis. That study helped put to rest the conventional wisdom that proteins actually rest at all.
This Nature study takes Kern's research to the next level, seeing the high-resolution structure of the hidden, high-energy state for the first time. For this success, high -resolution x-ray crystallography was further pushed by analyzing electron density data previously discarded as "noise" and by collecting data at ambient temperature. The protein of interest is human cyclophilin A, an enzyme that is highjacked by the HIV virus to aid its own replication.
But it was thanks to some clever protein design together with dynamic NMR spectroscopy that provided direct experimental evidence that the hidden structures in the high-energy state are in fact essential for catalysis. The researchers revealed what happens when proteins flip from the rare state to a major state in a process called interconversion. If this flip is fast, then the enzyme does its job fast, but if the flip is slow, as in the designer enzyme, then the enzyme operates slowly.
"People always focused on the chemistry—accelerating the reaction through catalyzing the chemical step of the substrates. What we've shown is that protein dynamics is as important as the chemical step," said Kern, a Howard Hughes Medical Institute Investigator. "Basically, all the steps need to be choreographed just right, like steps for a beautiful dancer. An enzyme can only function well with the perfect choreography of all the components."
Said Kern: "We now can show directly that the higher energy states are always there and that these hidden, rare states are absolutely essential for protein function."