Oct 11 2013
The microscopic technique, developed by researchers at Queen Mary University of London, represents a major advance for cell biologists as it will allow them to investigate structures deep inside the cell, such as viruses, bacteria and parts of the nucleus in depth.
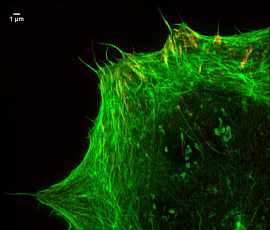 Focal adhesions and actin
Focal adhesions and actin
Recent advances in optical physics have made it possible to use fluorescent microscopy to study complex structures smaller than 200 nanometres (nm) – around 500 times smaller than the width of a human hair. These methodologies are called super-resolution microscopy.
The drawback of such techniques is that they can only produce very clear images of structures that are at the bottom of the cell. Since the nucleus – the cell’s ‘control centre’ – is in the middle of the cell and bacterial and viral infections can happen anywhere in the cell, this technique has considerable limitations for biologists.
This study shows how these issues have been overcome with a newly developed imaging system, making it possible to image structures as small as 80nm or less anywhere in the cell. The Spinning Disk Statistical Imaging (SDSI) system was developed by Dr Neveen Hosny, a bioengineer working with Professor Martin Knight in the School of Engineering and Materials Science and Dr Ann Wheeler, Head of Imaging at Queen Mary’s Blizard Institute.
Dr Ann Wheeler said: “The spinning disk microscope produces focused images at high speed because it has a disk with an array of tiny holes in it which remove the out of focus light. We have combined this microscope with new fluorescent probes, which switch between a bright and dark state rapidly. This system is now allowing us to see structures three times smaller than could usually be seen using standard light microscopes.
“We have been able to visualise chromatin, which is the protein structure that controls DNA expression and the nuclear envelope. We have also used the method to get images of focal adhesions – sub-cellular macromolecules which the cell uses to attach to its environment.
“Although it was previously possible to see these structures, our method provides a greater degree of detail. It also allows us to look at protein complexes which are smaller than 200nm in the nucleus, which hasn't been done before.”
The microscope is housed in its own room with a carefully controlled environment to miminise vibrations.
Professor Knight added: “Super resolution microscopy is a major step forward and we are looking forward to using this technology in a wide range of applications from stem cell behaviour to understanding arthritis or the development of nanomedicine.”
Dr Wheeler has worked with colleagues across Queen Mary to make the technique cost-effective and easy to use for scientists who are not experts in optical physics.
Dr Wheeler added: “We will be continuing to develop the technology to improve the fluorescent probes used for this technique and also applying it to cellular processes such as invasion in cancer.”
The development and an analysis of the SDSI system is published today (Wednesday 9 October) in the journal PLOS ONE.